Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells
- PMID: 25896322
- PMCID: PMC4503379
- DOI: 10.1038/nature14308
Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells
Abstract
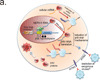
Endogenous retroviruses (ERVs) are remnants of ancient retroviral infections, and comprise nearly 8% of the human genome. The most recently acquired human ERV is HERVK(HML-2), which repeatedly infected the primate lineage both before and after the divergence of the human and chimpanzee common ancestor. Unlike most other human ERVs, HERVK retained multiple copies of intact open reading frames encoding retroviral proteins. However, HERVK is transcriptionally silenced by the host, with the exception of in certain pathological contexts such as germ-cell tumours, melanoma or human immunodeficiency virus (HIV) infection. Here we demonstrate that DNA hypomethylation at long terminal repeat elements representing the most recent genomic integrations, together with transactivation by OCT4 (also known as POU5F1), synergistically facilitate HERVK expression. Consequently, HERVK is transcribed during normal human embryogenesis, beginning with embryonic genome activation at the eight-cell stage, continuing through the emergence of epiblast cells in preimplantation blastocysts, and ceasing during human embryonic stem cell derivation from blastocyst outgrowths. Remarkably, we detected HERVK viral-like particles and Gag proteins in human blastocysts, indicating that early human development proceeds in the presence of retroviral products. We further show that overexpression of one such product, the HERVK accessory protein Rec, in a pluripotent cell line is sufficient to increase IFITM1 levels on the cell surface and inhibit viral infection, suggesting at least one mechanism through which HERVK can induce viral restriction pathways in early embryonic cells. Moreover, Rec directly binds a subset of cellular RNAs and modulates their ribosome occupancy, indicating that complex interactions between retroviral proteins and host factors can fine-tune pathways of early human development.
Figures













References
Additional references
-
- Chavez SL, Meneses JJ, Nguyen HN, Kim SK, Pera RAR. Characterization of Six New Human Embryonic Stem Cell Lines (HSF7, −8, −9, −10, −12, and −13) Derived Under Minimal-Animal Component Conditions. Stem Cells Dev. 2008;17:535–546. - PubMed
-
- Myers JWJEF., Jr . In: RNA Silencing. Carmichael GG, editor. Humana Press; 2005. pp. 93–196. at < http://link.springer.com/protocol/10.1385/1-59259-935-4%3A093>. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- DP2AI11219301/AI/NIAID NIH HHS/United States
- R01 GM112720/GM/NIGMS NIH HHS/United States
- U01 HL100397/HL/NHLBI NIH HHS/United States
- P01GM099130/GM/NIGMS NIH HHS/United States
- P50-HG007735/HG/NHGRI NIH HHS/United States
- DP2 AI112193/AI/NIAID NIH HHS/United States
- 1S10RR02678001/RR/NCRR NIH HHS/United States
- P01 GM099130/GM/NIGMS NIH HHS/United States
- P50 HG007735/HG/NHGRI NIH HHS/United States
- 1S10RR02933801/RR/NCRR NIH HHS/United States
- Howard Hughes Medical Institute/United States
- T32 HG000044/HG/NHGRI NIH HHS/United States
- 1F30CA189514-01/CA/NCI NIH HHS/United States
- F30 CA189514/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

