A STIM2 splice variant negatively regulates store-operated calcium entry
- PMID: 25896806
- PMCID: PMC4411291
- DOI: 10.1038/ncomms7899
A STIM2 splice variant negatively regulates store-operated calcium entry
Abstract
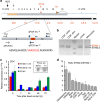
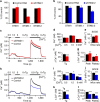
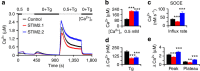
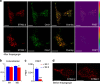
Cellular homeostasis relies upon precise regulation of Ca(2+) concentration. Stromal interaction molecule (STIM) proteins regulate store-operated calcium entry (SOCE) by sensing Ca(2+) concentration in the ER and forming oligomers to trigger Ca(2+) entry through plasma membrane-localized Orai1 channels. Here we characterize a STIM2 splice variant, STIM2.1, which retains an additional exon within the region encoding the channel-activating domain. Expression of STIM2.1 is ubiquitous but its abundance relative to the more common STIM2.2 variant is dependent upon cell type and highest in naive T cells. STIM2.1 knockdown increases SOCE in naive CD4(+) T cells, whereas knockdown of STIM2.2 decreases SOCE. Conversely, overexpression of STIM2.1, but not STIM2.2, decreases SOCE, indicating its inhibitory role. STIM2.1 interaction with Orai1 is impaired and prevents Orai1 activation, but STIM2.1 shows increased affinity towards calmodulin. Our results imply STIM2.1 as an additional player tuning Orai1 activation in vivo.
Figures




References
-
- Demaurex N. & Frieden M. Measurements of the free luminal ER Ca(2+) concentration with targeted ‘cameleon' fluorescent proteins. Cell Calcium 34, 109–119 (2003). - PubMed
-
- Prakriya M. E. Store-operated calcium channels. Curr. Top. Membr. 71, 1–273 (2013). - PubMed
-
- Soboloff J. et al. Orai1 and STIM reconstitute store-operated calcium channel function. J. Biol. Chem. 281, 20661–20665 (2006). - PubMed
-
- Zheng L., Stathopulos P. B., Li G. Y. & Ikura M. Biophysical characterization of the EF-hand and SAM domain containing Ca2+ sensory region of STIM1 and STIM2. Biochem. Biophys. Res. Commun. 369, 240–246 (2008). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

