Do larger genomes contain more diverse transposable elements?
- PMID: 25896861
- PMCID: PMC4438587
- DOI: 10.1186/s12862-015-0339-8
Do larger genomes contain more diverse transposable elements?
Abstract
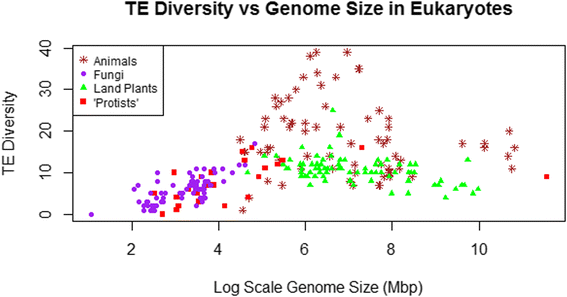
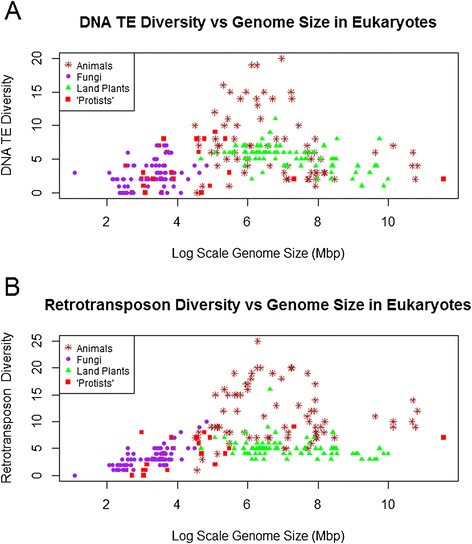
Background: The genomes of eukaryotes vary enormously in size, with much of this diversity driven by differences in the abundances of transposable elements (TEs). There is also substantial structural and phylogenetic diversity among TEs, such that they can be classified into distinct classes, superfamilies, and families. Possible relationships between TE diversity (and not just abundance) and genome size have not been investigated to date, though there are reasons to expect either a positive or a negative correlation. This study compares data from 257 species of animals, plants, fungi, and "protists" to determine whether TE diversity at the superfamily level is related to genome size.
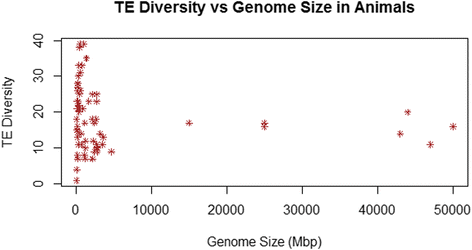
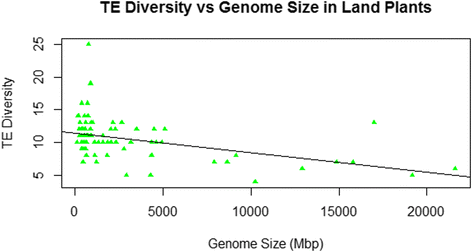
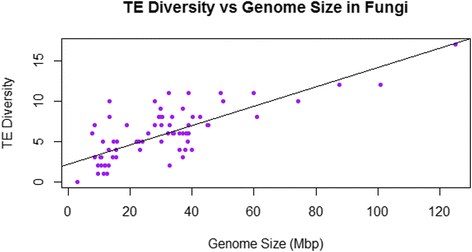
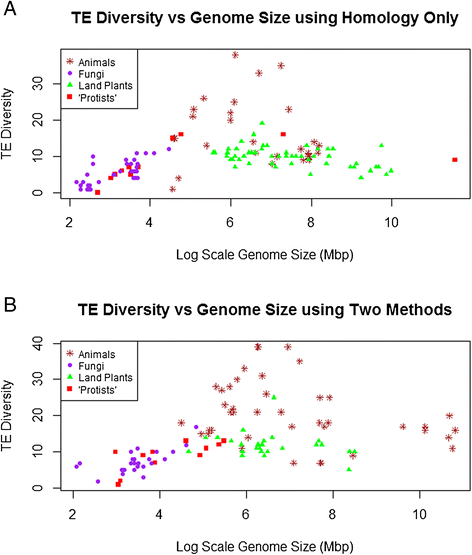
Results: No simple relationship was found between TE diversity and genome size. There is no significant correlation across all eukaryotes, but there is a positive correlation for genomes below 500 Mbp and a negative correlation among land plants. No relationships were found across animals or within vertebrates. Some TE superfamilies tend to be present across all major groups of eukaryotes, but there is considerable variance in TE diversity in different taxa.
Conclusions: Differences in genome size are thought to arise primarily through accumulation of TEs, but beyond a certain point (~500 Mbp), TE diversity does not increase with genome size. Several possible explanations for these complex patterns are discussed, and recommendations to facilitate future analyses are provided.
Figures
References
-
- Gregory TR, DeSalle R. Comparative genomics in eukaryotes. In: Gregory TR, editor. The Evolution of the Genome. San Diego: Elsevier; 2005. pp. 585–675.
-
- Pellicer J, Fay MF, Leitch IJ. The largest eukaryotic genome of them all? Bot J Linn Soc. 2010;164:10–15. doi: 10.1111/j.1095-8339.2010.01072.x. - DOI
-
- Gregory TR. Animal Genome Size Database. [http://www.genomesize.com]
-
- Bennett MD, Leitch IJ. Plant DNA C-values Database. [http://data.kew.org/cvalues/]
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

