Clinically Relevant Molecular Subtypes in Leiomyosarcoma
- PMID: 25896974
- PMCID: PMC4526352
- DOI: 10.1158/1078-0432.CCR-14-3141
Clinically Relevant Molecular Subtypes in Leiomyosarcoma
Abstract
Purpose: Leiomyosarcoma is a malignant neoplasm with smooth muscle differentiation. Little is known about its molecular heterogeneity and no targeted therapy currently exists for leiomyosarcoma. Recognition of different molecular subtypes is necessary to evaluate novel therapeutic options. In a previous study on 51 leiomyosarcomas, we identified three molecular subtypes in leiomyosarcoma. The current study was performed to determine whether the existence of these subtypes could be confirmed in independent cohorts.
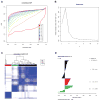
Experimental design: Ninety-nine cases of leiomyosarcoma were expression profiled with 3'end RNA-Sequencing (3SEQ). Consensus clustering was conducted to determine the optimal number of subtypes.
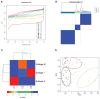
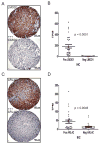
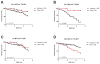
Results: We identified 3 leiomyosarcoma molecular subtypes and confirmed this finding by analyzing publically available data on 82 leiomyosarcoma from The Cancer Genome Atlas (TCGA). We identified two new formalin-fixed, paraffin-embedded tissue-compatible diagnostic immunohistochemical markers; LMOD1 for subtype I leiomyosarcoma and ARL4C for subtype II leiomyosarcoma. A leiomyosarcoma tissue microarray with known clinical outcome was used to show that subtype I leiomyosarcoma is associated with good outcome in extrauterine leiomyosarcoma while subtype II leiomyosarcoma is associated with poor prognosis in both uterine and extrauterine leiomyosarcoma. The leiomyosarcoma subtypes showed significant differences in expression levels for genes for which novel targeted therapies are being developed, suggesting that leiomyosarcoma subtypes may respond differentially to these targeted therapies.
Conclusions: We confirm the existence of 3 molecular subtypes in leiomyosarcoma using two independent datasets and show that the different molecular subtypes are associated with distinct clinical outcomes. The findings offer an opportunity for treating leiomyosarcoma in a subtype-specific targeted approach.
©2015 American Association for Cancer Research.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures
References
-
- Gustafson P, Willen H, Baldetorp B, Ferno M, Akerman M, Rydholm A. Soft tissue leiomyosarcoma. A population-based epidemiologic and prognostic study of 48 patients, including cellular DNA content. Cancer. 1992;70:114–9. - PubMed
-
- Toro JR, Travis LB, Wu HJ, Zhu K, Fletcher CD, Devesa SS. Incidence patterns of soft tissue sarcomas, regardless of primary site, in the surveillance, epidemiology and end results program, 1978–2001: An analysis of 26,758 cases. Int J Cancer. 2006;119:2922–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

