A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin
- PMID: 25897118
- PMCID: PMC4482077
- DOI: 10.1093/nar/gkv321
A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin
Abstract
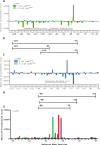
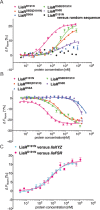
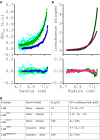
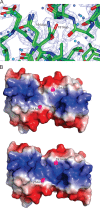
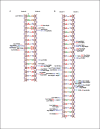
LiaR is a 'master regulator' of the cell envelope stress response in enterococci and many other Gram-positive organisms. Mutations to liaR can lead to antibiotic resistance to a variety of antibiotics including the cyclic lipopeptide daptomycin. LiaR is phosphorylated in response to membrane stress to regulate downstream target operons. Using DNA footprinting of the regions upstream of the liaXYZ and liaFSR operons we show that LiaR binds an extended stretch of DNA that extends beyond the proposed canonical consensus sequence suggesting a more complex level of regulatory control of target operons. We go on to determine the biochemical and structural basis for increased resistance to daptomycin by the adaptive mutation to LiaR (D191N) first identified from the pathogen Enterococcus faecalis S613. LiaR(D191N) increases oligomerization of LiaR to form a constitutively activated tetramer that has high affinity for DNA even in the absence of phosphorylation leading to increased resistance. Crystal structures of the LiaR DNA binding domain complexed to the putative consensus sequence as well as an adjoining secondary sequence show that upon binding, LiaR induces DNA bending that is consistent with increased recruitment of RNA polymerase to the transcription start site and upregulation of target operons.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


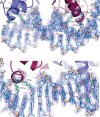

Similar articles
-
Two Mutations Commonly Associated with Daptomycin Resistance in Enterococcus faecium LiaST120A and LiaRW73C Appear To Function Epistatically in LiaFSR Signaling.Biochemistry. 2018 Dec 11;57(49):6797-6805. doi: 10.1021/acs.biochem.8b01072. Epub 2018 Nov 27. Biochemistry. 2018. PMID: 30403130 Free PMC article.
-
An Adaptive Mutation in Enterococcus faecium LiaR Associated with Antimicrobial Peptide Resistance Mimics Phosphorylation and Stabilizes LiaR in an Activated State.J Mol Biol. 2016 Nov 6;428(22):4503-4519. doi: 10.1016/j.jmb.2016.09.016. Epub 2016 Sep 23. J Mol Biol. 2016. PMID: 27670715 Free PMC article.
-
A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis.J Infect Dis. 2015 Apr 15;211(8):1317-25. doi: 10.1093/infdis/jiu602. Epub 2014 Oct 31. J Infect Dis. 2015. PMID: 25362197 Free PMC article.
-
Mechanisms of drug resistance: daptomycin resistance.Ann N Y Acad Sci. 2015 Sep;1354:32-53. doi: 10.1111/nyas.12948. Epub 2015 Oct 23. Ann N Y Acad Sci. 2015. PMID: 26495887 Free PMC article. Review.
-
Daptomycin: a lipopeptide antibiotic for the treatment of serious Gram-positive infections.J Antimicrob Chemother. 2005 Mar;55(3):283-8. doi: 10.1093/jac/dkh546. Epub 2005 Feb 10. J Antimicrob Chemother. 2005. PMID: 15705644 Review.
Cited by
-
Two Mutations Commonly Associated with Daptomycin Resistance in Enterococcus faecium LiaST120A and LiaRW73C Appear To Function Epistatically in LiaFSR Signaling.Biochemistry. 2018 Dec 11;57(49):6797-6805. doi: 10.1021/acs.biochem.8b01072. Epub 2018 Nov 27. Biochemistry. 2018. PMID: 30403130 Free PMC article.
-
LiaR-dependent gene expression contributes to antimicrobial responses in group A Streptococcus.Antimicrob Agents Chemother. 2024 Dec 5;68(12):e0049624. doi: 10.1128/aac.00496-24. Epub 2024 Nov 13. Antimicrob Agents Chemother. 2024. PMID: 39535201 Free PMC article.
-
The LiaFSR-LiaX System Mediates Resistance of Enterococcus faecium to Peptide Antibiotics and to Aureocin A53- and Enterocin L50-Like Bacteriocins.Microbiol Spectr. 2023 Jun 15;11(3):e0034323. doi: 10.1128/spectrum.00343-23. Epub 2023 May 23. Microbiol Spectr. 2023. PMID: 37219451 Free PMC article.
-
Molecular Bases Determining Daptomycin Resistance-Mediated Resensitization to β-Lactams (Seesaw Effect) in Methicillin-Resistant Staphylococcus aureus.Antimicrob Agents Chemother. 2016 Dec 27;61(1):e01634-16. doi: 10.1128/AAC.01634-16. Print 2017 Jan. Antimicrob Agents Chemother. 2016. PMID: 27795377 Free PMC article.
-
An Adaptive Mutation in Enterococcus faecium LiaR Associated with Antimicrobial Peptide Resistance Mimics Phosphorylation and Stabilizes LiaR in an Activated State.J Mol Biol. 2016 Nov 6;428(22):4503-4519. doi: 10.1016/j.jmb.2016.09.016. Epub 2016 Sep 23. J Mol Biol. 2016. PMID: 27670715 Free PMC article.
References
-
- Boucher H.W., Talbot G.H., Bradley J.S., Edwards J.E., Gilbert D., Rice L.B., Scheld M., Spellberg B., Bartlett J. Bad bugs, no drugs: no ESKAPE! an update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48:1–12. - PubMed
-
- Rice L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 2008;197:1079–1081. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

