miRiadne: a web tool for consistent integration of miRNA nomenclature
- PMID: 25897123
- PMCID: PMC4489305
- DOI: 10.1093/nar/gkv381
miRiadne: a web tool for consistent integration of miRNA nomenclature
Abstract
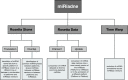
The miRBase is the official miRNA repository which keeps the annotation updated on newly discovered miRNAs: it is also used as a reference for the design of miRNA profiling platforms. Nomenclature ambiguities generated by loosely updated platforms and design errors lead to incompatibilities among platforms, even from the same vendor. Published miRNA lists are thus generated with different profiling platforms that refer to diverse and not updated annotations. This greatly compromises searches, comparisons and analyses that rely on miRNA names only without taking into account the mature sequences, which is particularly critic when such analyses are carried over automatically. In this paper we introduce miRiadne, a web tool to harmonize miRNA nomenclature, which takes into account the original miRBase versions from 10 up to 21, and annotations of 40 common profiling platforms from nine brands that we manually curated. miRiadne uses the miRNA mature sequence to link miRBase versions and/or platforms to prevent nomenclature ambiguities. miRiadne was designed to simplify and support biologists and bioinformaticians in re-annotating their own miRNA lists and/or data sets. As Ariadne helped Theseus in escaping the mythological maze, miRiadne will help the miRNA researcher in escaping the nomenclature maze. miRiadne is freely accessible from the URL http://www.miriadne.org.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Zhang B., Wang Q., Pan X. MicroRNAs and their regulatory roles in animals and plants. J. Cell. Physiol. 2007;210:279–289. - PubMed
-
- Nelson K.M., Weiss G.J. MicroRNAs and cancer: past, present, and potential future. Mol. Cancer Ther. 2008;7:3655–3660. - PubMed
-
- Pagani M., Rossetti G., Panzeri I., de Candia P., Bonnal R.J.P., Rossi R.L., Geginat J., Abrignani S. Role of microRNAs and long-non-coding RNAs in CD4(+) T-cell differentiation. Immunol. Rev. 2013;253:82–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

