The amino-terminal tails of histones H2A and H3 coordinate efficient base excision repair, DNA damage signaling and postreplication repair in Saccharomyces cerevisiae
- PMID: 25897129
- PMCID: PMC4446432
- DOI: 10.1093/nar/gkv372
The amino-terminal tails of histones H2A and H3 coordinate efficient base excision repair, DNA damage signaling and postreplication repair in Saccharomyces cerevisiae
Abstract
Histone amino-terminal tails (N-tails) are required for cellular resistance to DNA damaging agents; therefore, we examined the role of histone N-tails in regulating DNA damage response pathways in Saccharomyces cerevisiae. Combinatorial deletions reveal that the H2A and H3 N-tails are important for the removal of MMS-induced DNA lesions due to their role in regulating the basal and MMS-induced expression of DNA glycosylase Mag1. Furthermore, overexpression of Mag1 in a mutant lacking the H2A and H3 N-tails rescues base excision repair (BER) activity but not MMS sensitivity. We further show that the H3 N-tail functions in the Rad9/Rad53 DNA damage signaling pathway, but this function does not appear to be the primary cause of MMS sensitivity of the double tailless mutants. Instead, epistasis analyses demonstrate that the tailless H2A/H3 phenotypes are in the RAD18 epistasis group, which regulates postreplication repair. We observed increased levels of ubiquitylated PCNA and significantly lower mutation frequency in the tailless H2A/H3 mutant, indicating a defect in postreplication repair. In summary, our data identify novel roles of the histone H2A and H3 N-tails in (i) regulating the expression of a critical BER enzyme (Mag1), (ii) supporting efficient DNA damage signaling and (iii) facilitating postreplication repair.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


 , WT;
, WT;  , tH2A:tH3; and
, tH2A:tH3; and  , mag1Δ). The % of methylpurines removed at each time (or % repair) is shown in the graph as mean ± standard deviation of at least three independent experiments. The initial methylpurines/kb are listed in the table. (B) Repair was analyzed at the GAL10 locus by Southern blotting using locus-specific probes. Graph depicts mean ± standard deviation of at least three independent experiments. (C) High resolution DNA damage mapping was performed on the NTS of RPB2 for WT and tH2A:tH3 cells. The top band on the sequencing gel represents the full-length band, while bands that migrate faster represent methylpurine damage sites that have been cleaved by AAG and APE1 digestion. The arrow on the left indicates the transcription start site and the ovals on the right indicate the most prominent nucleosome positions along RPB2 as mapped by Jiang et al. (34). The plot is aligned so that each point is indicative of the t0.5 (time to repair 50% of lesions) for its respective band. The smoothed lines were determined by averaging the t0.5 of 41 nucleotides surround each nucleotide site, where the 41 nucleotides encompass the nucleotide position and 20 nucleotides flanking that nucleotide in each direction.
, mag1Δ). The % of methylpurines removed at each time (or % repair) is shown in the graph as mean ± standard deviation of at least three independent experiments. The initial methylpurines/kb are listed in the table. (B) Repair was analyzed at the GAL10 locus by Southern blotting using locus-specific probes. Graph depicts mean ± standard deviation of at least three independent experiments. (C) High resolution DNA damage mapping was performed on the NTS of RPB2 for WT and tH2A:tH3 cells. The top band on the sequencing gel represents the full-length band, while bands that migrate faster represent methylpurine damage sites that have been cleaved by AAG and APE1 digestion. The arrow on the left indicates the transcription start site and the ovals on the right indicate the most prominent nucleosome positions along RPB2 as mapped by Jiang et al. (34). The plot is aligned so that each point is indicative of the t0.5 (time to repair 50% of lesions) for its respective band. The smoothed lines were determined by averaging the t0.5 of 41 nucleotides surround each nucleotide site, where the 41 nucleotides encompass the nucleotide position and 20 nucleotides flanking that nucleotide in each direction.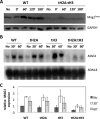


Similar articles
-
Nucleosomes Regulate Base Excision Repair in Chromatin.Mutat Res Rev Mutat Res. 2019 Apr-Jun;780:29-36. doi: 10.1016/j.mrrev.2017.10.002. Epub 2017 Nov 7. Mutat Res Rev Mutat Res. 2019. PMID: 31388331 Free PMC article. Review.
-
Methylation of histone H3 lysine-79 by Dot1p plays multiple roles in the response to UV damage in Saccharomyces cerevisiae.DNA Repair (Amst). 2007 Mar 1;6(3):383-95. doi: 10.1016/j.dnarep.2006.12.010. Epub 2007 Jan 30. DNA Repair (Amst). 2007. PMID: 17267293
-
Genetic analysis of Saccharomyces cerevisiae H2A serine 129 mutant suggests a functional relationship between H2A and the sister-chromatid cohesion partners Csm3-Tof1 for the repair of topoisomerase I-induced DNA damage.Genetics. 2006 Jan;172(1):67-76. doi: 10.1534/genetics.105.046128. Epub 2005 Oct 11. Genetics. 2006. PMID: 16219777 Free PMC article.
-
Previously uncharacterized amino acid residues in histone H3 and H4 mutants with roles in DNA damage repair response and cellular aging.FEBS J. 2019 Mar;286(6):1154-1173. doi: 10.1111/febs.14723. Epub 2018 Dec 28. FEBS J. 2019. PMID: 30536627
-
The contribution of the budding yeast histone H2A C-terminal tail to DNA-damage responses.Biochem Soc Trans. 2007 Dec;35(Pt 6):1519-24. doi: 10.1042/BST0351519. Biochem Soc Trans. 2007. PMID: 18031258 Review.
Cited by
-
NuA4 acetyltransferase is required for efficient nucleotide excision repair in yeast.DNA Repair (Amst). 2019 Jan;73:91-98. doi: 10.1016/j.dnarep.2018.11.006. Epub 2018 Nov 14. DNA Repair (Amst). 2019. PMID: 30473425 Free PMC article.
-
Synergistic effects of H3 and H4 nucleosome tails on structure and dynamics of a lesion-containing DNA: Binding of a displaced lesion partner base to the H3 tail for GG-NER recognition.DNA Repair (Amst). 2018 May;65:73-78. doi: 10.1016/j.dnarep.2018.02.009. Epub 2018 Mar 8. DNA Repair (Amst). 2018. PMID: 29631253 Free PMC article.
-
Nucleosomes Regulate Base Excision Repair in Chromatin.Mutat Res Rev Mutat Res. 2019 Apr-Jun;780:29-36. doi: 10.1016/j.mrrev.2017.10.002. Epub 2017 Nov 7. Mutat Res Rev Mutat Res. 2019. PMID: 31388331 Free PMC article. Review.
-
Genome Instability Is Promoted by the Chromatin-Binding Protein Spn1 in Saccharomyces cerevisiae.Genetics. 2018 Dec;210(4):1227-1237. doi: 10.1534/genetics.118.301600. Epub 2018 Oct 9. Genetics. 2018. PMID: 30301740 Free PMC article.
-
Direct assessment of histone function using histone replacement.Trends Biochem Sci. 2023 Jan;48(1):53-70. doi: 10.1016/j.tibs.2022.06.010. Epub 2022 Jul 16. Trends Biochem Sci. 2023. PMID: 35853806 Free PMC article. Review.
References
-
- Friedberg E.C., Walker G.C., Siede W., Wood R.D., Schultz R.A., Ellenberger T. DNA Repair and Mutagenesis. 2nd edn. Washington, D.C: ASM Press; 2006.
-
- Kondo T., Wakayama T., Naiki T., Matsumoto K., Sugimoto K. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science. 2001;294:867–870. - PubMed
-
- Pellicioli A., Foiani M. Signal transduction: how rad53 kinase is activated. Curr. Biol. 2005;15:R769–R771. - PubMed
-
- Branzei D., Foiani M. The Rad53 signal transduction pathway: replication fork stabilization, DNA repair, and adaptation. Exp. Cell Res. 2006;312:2654–2659. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

