Synthetic viability genomic screening defines Sae2 function in DNA repair
- PMID: 25899817
- PMCID: PMC4474527
- DOI: 10.15252/embj.201590973
Synthetic viability genomic screening defines Sae2 function in DNA repair
Abstract
DNA double-strand break (DSB) repair by homologous recombination (HR) requires 3' single-stranded DNA (ssDNA) generation by 5' DNA-end resection. During meiosis, yeast Sae2 cooperates with the nuclease Mre11 to remove covalently bound Spo11 from DSB termini, allowing resection and HR to ensue. Mitotic roles of Sae2 and Mre11 nuclease have remained enigmatic, however, since cells lacking these display modest resection defects but marked DNA damage hypersensitivities. By combining classic genetic suppressor screening with high-throughput DNA sequencing, we identify Mre11 mutations that strongly suppress DNA damage sensitivities of sae2∆ cells. By assessing the impacts of these mutations at the cellular, biochemical and structural levels, we propose that, in addition to promoting resection, a crucial role for Sae2 and Mre11 nuclease activity in mitotic DSB repair is to facilitate the removal of Mre11 from ssDNA associated with DSB ends. Thus, without Sae2 or Mre11 nuclease activity, Mre11 bound to partly processed DSBs impairs strand invasion and HR.
Keywords: Mre11; Sae2; suppressor screening; synthetic viability; whole‐genome sequencing.
© 2015 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
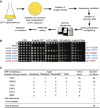
Outline of the screening approach that was used to identify suppressors of sae2Δ camptothecin (CPT) hypersensitivity.
Validation of the suppression phenotypes; a subset (sup25–sup30) of the suppressors recovered from the screening is shown along with mutations identified in each clone.
Summary of the results of the synthetic viability genomic screening (SVGS) for sae2Δ camptothecin (CPT) hypersensitivity. The ORF and the type of mutation are reported together with the number of times each ORF was found mutated and the number of clones in which each ORF was putatively driving the resistance.

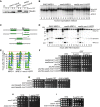
A Alignment of Mre11 region containing H37 in fungal species; secondary structure prediction is shown above.
B Western blot with anti-Mre11 antibody on protein extracts prepared from the indicated strains shows that mre11-H37R and mre11-H37Y mutations do not alter Mre11 protein levels (* indicate cross-reacting proteins).
C sup28 and sup29 suppression is rescued by expressing wild-type (wt) Mre11.
D mre11-H37R and mre11-H37Y suppress sae2Δ DNA damage hypersensitivity.
E, F mre11-H37Y suppresses DNA damage hypersensitivities of sae2MT (sae2-2,5,6,8,9) and sae2-S267A cells. CPT, camptothecin; Phleo, phleomycin.
G mre11-H37A does not suppress sae2Δ.
A mre11-H37R suppresses sae2Δ checkpoint hyperactivation.
B mre11-H37R does not rescue sae2Δ meiotic DSB processing defect.
C Outline of DSB repair by single-strand annealing (SSA).
D mre11-H37R does not rescue the SSA repair defect of sae2Δ strains.
E mre11-H37R does not rescue sae2Δ-dependent cell cycle arrest caused by DSB induction.
F, G Exo1 and Ku are not required for mre11-H37R-mediated suppression of sae2Δ hypersensitivity.
H mre11-H37R does not suppress xrs2Δ camptothecin (CPT) hypersensitivity.
I mre11-H37R suppresses rad50S CPT hypersensitivity.

A Mre11 and Mre11H37R were purified to homogeneity from yeast cultures.
B 3′ exonuclease activity assay on Mre11 and Mre11H37R leading to release of a labelled single nucleotide, as indicated.
C, D Electrophoretic mobility shift assays on Mre11, Mre11H37R and Mre11H37A with dsDNA (C) or ssDNA (D).
E Quantification of mre11-H37R suppression of sae2Δ cell DNA damage hypersensitivity. Overnight grown cultures of the indicated strains were diluted and plated on medium containing the indicated doses of CPT. Colony growth was scored 3–6 days later. Averages and standard deviations are shown for each point.
F Intragenic suppression of CPT hypersensitivity of mre11-nd (mre11-H125N) by mre11-H37R. Overnight grown cultures of the indicated strains were treated as in (E). Dotted lines represent data from (E). Averages and standard deviations are shown for each point.
G Mre11 nuclease activity is not required for mre11-H37R-mediated suppression of sae2Δ CPT hypersensitivity. Overnight grown cultures of the indicated strains were treated as in (E). The dotted lines represent data from (E). Averages and standard deviations are shown for each point.
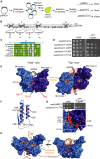
Outline of the plasmid mutagenesis approach to identify new mre11SUPsae2Δ alleles. LOF: loss-of-function alleles. SUP: suppressor alleles.
Mre11 with shaded boxes and blue shapes indicating phosphoesterase motifs and secondary structures, respectively; additional mre11SUPsae2Δ mutations recovered from the screening are indicated.
Fungal alignment and secondary structure prediction of the region of Mre11 containing Q70.
mre11-Q70R, mre11-L89V and mre11-P110L alleles recovered from plasmid mutagenesis screening suppress sae2Δ hypersensitivity to camptothecin.
Structural prediction of S. cerevisiae Mre11 residues 1–414, obtained by homology modelling using the corresponding S. pombe and human structures. The water-accessible surface of the two monomers is shown in different shades of blue. Red: residues whose mutation suppresses sae2Δ DNA damage hypersensitivity. Orange: residues whose mutation abrogates Mre11 nuclease activity.
Model of Mre11 tertiary structure (residues 1–100). Residues are colour-coded as in (E).
Top: mre11-L77R suppresses the DNA damage hypersensitivity of sae2Δ cells. Bottom: localisation of mre11SUPsae2Δ suppressors on the molecular model of the Mre11 dimer. The two Mre11 monomers are shown in different shades of blue, and the proposed path of bound ssDNA is indicated by the orange filament.
Model in which the two DNA filaments of the two DSB ends melt when binding to Mre11; the 5′ ends being channelled towards the active site and the 3′ end being channelled towards the Mre11SUPsae2Δ region.

IR-induced Mre11H37R foci (IRIF) persist for shorter times than Mre11-wt IRIF in exponentially growing sae2Δ cells (average and standard deviations from two or more independent experiments).
Effects of sae2Δ and mre11-H37R on Mre11 IRIF persistence still occur when Rad51 is absent, revealing that Mre11 IRIF persistence causes defective HR (average and standard deviation from two independent experiments).
mre11-H37R suppresses Mre11 IRIF persistence in exponentially growing rad50S cells (average and standard deviation from two independent experiments).
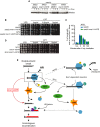
mre11-H37R suppresses Tel1 hyperactivation induced by Mre11 IRIF persistence in sae2Δ cells.
Deletion of TEL1 weakens the suppression of the sensitivity of a sae2Δ strain mediated by mre11-H37R.
Deletion of TEL1 reduces the hyperaccumulation of Mre11 to IRIF and impairs the suppression of their persistence mediated by mre11-H37R (average and standard deviation from two independent experiments).
mre11-H37R suppresses the sensitivity to CPT of a tel1Δ strain.
Model for the role of MRX, Sae2 and Tel1 in response to DSBs.
References
-
- Benkert P, Tosatto SCE, Schomburg D. QMEAN: a comprehensive scoring function for model quality assessment. Proteins. 2008;71:261–277. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 11224/CRUK_/Cancer Research UK/United Kingdom
- K99 ES021441/ES/NIEHS NIH HHS/United States
- C6/A11224/CRUK_/Cancer Research UK/United Kingdom
- 101126/Z/13/Z/WT_/Wellcome Trust/United Kingdom
- C6946/A14492/CRUK_/Cancer Research UK/United Kingdom
- 101126/WT_/Wellcome Trust/United Kingdom
- 13031/CRUK_/Cancer Research UK/United Kingdom
- R01 ES007061/ES/NIEHS NIH HHS/United States
- WT092096/WT_/Wellcome Trust/United Kingdom
- R01ES007061/ES/NIEHS NIH HHS/United States
- WT098051/WT_/Wellcome Trust/United Kingdom
- 18796/CRUK_/Cancer Research UK/United Kingdom
- K99ES021441/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

