BioID-based Identification of Skp Cullin F-box (SCF)β-TrCP1/2 E3 Ligase Substrates
- PMID: 25900982
- PMCID: PMC4587326
- DOI: 10.1074/mcp.M114.045658
BioID-based Identification of Skp Cullin F-box (SCF)β-TrCP1/2 E3 Ligase Substrates
Abstract
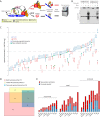
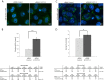
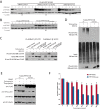
The identification of ubiquitin E3 ligase substrates has been challenging, due in part to low-affinity, transient interactions, the rapid degradation of targets and the inability to identify proteins from poorly soluble cellular compartments. SCF(β-TrCP1) and SCF(β-TrCP2) are well-studied ubiquitin E3 ligases that target substrates for proteasomal degradation, and play important roles in Wnt, Hippo, and NFκB signaling. Combining 26S proteasome inhibitor (MG132) treatment with proximity-dependent biotin labeling (BioID) and semiquantitative mass spectrometry, here we identify SCF(β-TrCP1/2) interacting partners. Based on their enrichment in the presence of MG132, our data identify over 50 new putative SCF(β-TrCP1/2) substrates. We validate 12 of these new substrates and reveal previously unsuspected roles for β-TrCP in the maintenance of nuclear membrane integrity, processing (P)-body turnover and translational control. Together, our data suggest that β-TrCP is an important hub in the cellular stress response. The technique presented here represents a complementary approach to more standard IP-MS methods and should be broadly applicable for the identification of substrates for many ubiquitin E3 ligases.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Varshavsky A. (2012) The ubiquitin system, an immense realm. Annu. Rev. Biochem. 81, 167–176 - PubMed
-
- Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 - PubMed
-
- Jiang Y. H., Beaudet A. L. (2004) Human disorders of ubiquitination and proteasomal degradation. Curr. Opin. Pediar. 16, 419–426 - PubMed
-
- Persaud A., Rotin D. (2011) Use of proteome arrays to globally identify substrates for E3 ubiquitin ligases. Methods Mol. Biol. 759, 215–224 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

