Identification of Regulatory and Cargo Proteins of Endosomal and Secretory Pathways in Arabidopsis thaliana by Proteomic Dissection
- PMID: 25900983
- PMCID: PMC4587325
- DOI: 10.1074/mcp.M115.050286
Identification of Regulatory and Cargo Proteins of Endosomal and Secretory Pathways in Arabidopsis thaliana by Proteomic Dissection
Abstract
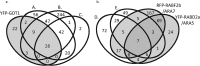
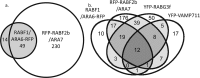
The cell's endomembranes comprise an intricate, highly dynamic and well-organized system. In plants, the proteins that regulate function of the various endomembrane compartments and their cargo remain largely unknown. Our aim was to dissect subcellular trafficking routes by enriching for partially overlapping subpopulations of endosomal proteomes associated with endomembrane markers. We selected RABD2a/ARA5, RABF2b/ARA7, RABF1/ARA6, and RABG3f as markers for combinations of the Golgi, trans-Golgi network (TGN), early endosomes (EE), secretory vesicles, late endosomes (LE), multivesicular bodies (MVB), and the tonoplast. As comparisons we used Golgi transport 1 (GOT1), which localizes to the Golgi, clathrin light chain 2 (CLC2) labeling clathrin-coated vesicles and pits and the vesicle-associated membrane protein 711 (VAMP711) present at the tonoplast. We developed an easy-to-use method by refining published protocols based on affinity purification of fluorescent fusion constructs to these seven subcellular marker proteins in Arabidopsis thaliana seedlings. We present a total of 433 proteins, only five of which were shared among all enrichments, while many proteins were common between endomembrane compartments of the same trafficking route. Approximately half, 251 proteins, were assigned to one enrichment only. Our dataset contains known regulators of endosome functions including small GTPases, SNAREs, and tethering complexes. We identify known cargo proteins such as PIN3, PEN3, CESA, and the recently defined TPLATE complex. The subcellular localization of two GTPase regulators predicted from our enrichments was validated using live-cell imaging. This is the first proteomic dataset to discriminate between such highly overlapping endomembrane compartments in plants and can be used as a general proteomic resource to predict the localization of proteins and identify the components of regulatory complexes and provides a useful tool for the identification of new protein markers of the endomembrane system.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

