TP53 loss creates therapeutic vulnerability in colorectal cancer
- PMID: 25901683
- PMCID: PMC4417759
- DOI: 10.1038/nature14418
TP53 loss creates therapeutic vulnerability in colorectal cancer
Erratum in
-
Author Correction: TP53 loss creates therapeutic vulnerability in colorectal cancer.Nature. 2021 Sep;597(7875):E6. doi: 10.1038/s41586-021-03664-3. Nature. 2021. PMID: 34433970 No abstract available.
Abstract
TP53, a well-known tumour suppressor gene that encodes p53, is frequently inactivated by mutation or deletion in most human tumours. A tremendous effort has been made to restore p53 activity in cancer therapies. However, no effective p53-based therapy has been successfully translated into clinical cancer treatment owing to the complexity of p53 signalling. Here we demonstrate that genomic deletion of TP53 frequently encompasses essential neighbouring genes, rendering cancer cells with hemizygous TP53 deletion vulnerable to further suppression of such genes. POLR2A is identified as such a gene that is almost always co-deleted with TP53 in human cancers. It encodes the largest and catalytic subunit of the RNA polymerase II complex, which is specifically inhibited by α-amanitin. Our analysis of The Cancer Genome Atlas (TCGA) and Cancer Cell Line Encyclopedia (CCLE) databases reveals that POLR2A expression levels are tightly correlated with its gene copy numbers in human colorectal cancer. Suppression of POLR2A with α-amanitin or small interfering RNAs selectively inhibits the proliferation, survival and tumorigenic potential of colorectal cancer cells with hemizygous TP53 loss in a p53-independent manner. Previous clinical applications of α-amanitin have been limited owing to its liver toxicity. However, we found that α-amanitin-based antibody-drug conjugates are highly effective therapeutic agents with reduced toxicity. Here we show that low doses of α-amanitin-conjugated anti-epithelial cell adhesion molecule (EpCAM) antibody lead to complete tumour regression in mouse models of human colorectal cancer with hemizygous deletion of POLR2A. We anticipate that inhibiting POLR2A will be a new therapeutic approach for human cancers containing such common genomic alterations.
Conflict of interest statement
J. Anderl and A. Pahl are employees of Heidelberg Pharma GmbH. The other authors declare no conflict of interest.
Figures







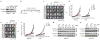

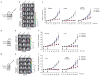




Comment in
-
Cancer: An essential passenger with p53.Nature. 2015 Apr 30;520(7549):626-7. doi: 10.1038/nature14390. Epub 2015 Apr 22. Nature. 2015. PMID: 25901678 No abstract available.
-
Colorectal cancer: A circuitous way to target p53.Nat Rev Cancer. 2015 Jun;15(6):318-9. doi: 10.1038/nrc3962. Epub 2015 May 14. Nat Rev Cancer. 2015. PMID: 25971675 No abstract available.
-
Colorectal cancer: POLR2A deletion with TP53 opens a window of opportunity for therapy.Nat Rev Clin Oncol. 2015 Jul;12(7):374. doi: 10.1038/nrclinonc.2015.94. Epub 2015 May 19. Nat Rev Clin Oncol. 2015. PMID: 25985938 No abstract available.
References
-
- Petitjean A, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28(6):622–629. - PubMed
-
- Vazquez A, et al. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7(12):979–987. - PubMed
-
- Cheok CF, et al. Translating p53 into the clinic. Nat Rev Clin Oncol. 2011;8(1):25–37. - PubMed
-
- Chene P. Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat Rev Cancer. 2003;3(2):102–109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

