The colibactin warhead crosslinks DNA
- PMID: 25901819
- PMCID: PMC4499846
- DOI: 10.1038/nchem.2221
The colibactin warhead crosslinks DNA
Abstract
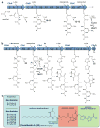
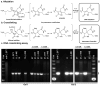
Members of the human microbiota are increasingly being correlated to human health and disease states, but the majority of the underlying microbial metabolites that regulate host-microbe interactions remain largely unexplored. Select strains of Escherichia coli present in the human colon have been linked to the initiation of inflammation-induced colorectal cancer through an unknown small-molecule-mediated process. The responsible non-ribosomal peptide-polyketide hybrid pathway encodes 'colibactin', which belongs to a largely uncharacterized family of small molecules. Genotoxic small molecules from this pathway that are capable of initiating cancer formation have remained elusive due to their high instability. Guided by metabolomic analyses, here we employ a combination of NMR spectroscopy and bioinformatics-guided isotopic labelling studies to characterize the colibactin warhead, an unprecedented substituted spirobicyclic structure. The warhead crosslinks duplex DNA in vitro, providing direct experimental evidence for colibactin's DNA-damaging activity. The data support unexpected models for both colibactin biosynthesis and its mode of action.
Conflict of interest statement
The authors declare no competing financial interests
Figures


Comment in
-
Natural products: Hunting microbial metabolites.Nat Chem. 2015 May;7(5):375-6. doi: 10.1038/nchem.2252. Nat Chem. 2015. PMID: 25901811 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

