Re-replication of a centromere induces chromosomal instability and aneuploidy
- PMID: 25901968
- PMCID: PMC4406714
- DOI: 10.1371/journal.pgen.1005039
Re-replication of a centromere induces chromosomal instability and aneuploidy
Abstract
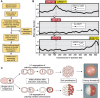
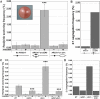
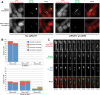
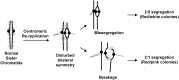
The faithful inheritance of chromosomes during cell division requires their precise replication and segregation. Numerous mechanisms ensure that each of these fundamental cell cycle events is performed with a high degree of fidelity. The fidelity of chromosomal replication is maintained in part by re-replication controls that ensure there are no more than two copies of every genomic segment to distribute to the two daughter cells. This control is enforced by inhibiting replication initiation proteins from reinitiating replication origins within a single cell cycle. Here we show in Saccharomyces cerevisiae that re-replication control is important for the fidelity of chromosome segregation. In particular, we demonstrate that transient re-replication of centromeric DNA due to disruption of re-replication control greatly induces aneuploidy of the re-replicated chromosome. Some of this aneuploidy arises from missegregation of both sister chromatids to one daughter cell. Aneuploidy can also arise from the generation of an extra sister chromatid via homologous recombination, suggesting that centromeric re-replication can trigger breakage and repair events that expand chromosome number without causing chromosomal rearrangements. Thus, we have identified a potential new non-mitotic source of aneuploidy that can arise from a defect in re-replication control. Given the emerging connections between the deregulation of replication initiation proteins and oncogenesis, this finding may be relevant to the aneuploidy that is prevalent in cancer.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Arias EE, Walter JC (2007) Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 497–518. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

