Capturing nature's diversity
- PMID: 25902039
- PMCID: PMC4406718
- DOI: 10.1371/journal.pone.0120942
Capturing nature's diversity
Abstract
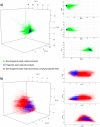
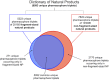
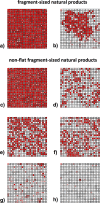
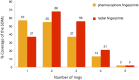
Natural products are universally recognized to contribute valuable chemical diversity to the design of molecular screening libraries. The analysis undertaken in this work, provides a foundation for the generation of fragment screening libraries that capture the diverse range of molecular recognition building blocks embedded within natural products. Physicochemical properties were used to select fragment-sized natural products from a database of known natural products (Dictionary of Natural Products). PCA analysis was used to illustrate the positioning of the fragment subset within the property space of the non-fragment sized natural products in the dataset. Structural diversity was analysed by three distinct methods: atom function analysis, using pharmacophore fingerprints, atom type analysis, using radial fingerprints, and scaffold analysis. Small pharmacophore triplets, representing the range of chemical features present in natural products that are capable of engaging in molecular interactions with small, contiguous areas of protein binding surfaces, were analysed. We demonstrate that fragment-sized natural products capture more than half of the small pharmacophore triplet diversity observed in non fragment-sized natural product datasets. Atom type analysis using radial fingerprints was represented by a self-organizing map. We examined the structural diversity of non-flat fragment-sized natural product scaffolds, rich in sp3 configured centres. From these results we demonstrate that 2-ring fragment-sized natural products effectively balance the opposing characteristics of minimal complexity and broad structural diversity when compared to the larger, more complex fragment-like natural products. These naturally-derived fragments could be used as the starting point for the generation of a highly diverse library with the scope for further medicinal chemistry elaboration due to their minimal structural complexity. This study highlights the possibility to capture a high proportion of the individual molecular interaction motifs embedded within natural products using a fragment screening library spanning 422 structural clusters and comprised of approximately 2800 natural products.
Conflict of interest statement
Figures

References
-
- Feher M, Schmidt JM (2002) Property Distributions: Differences between Drugs, Natural Products, and Molecules from Combinatorial Chemistry. J Chem Inf Comput Sci 43: 218–227. - PubMed
-
- Grabowski K, Schneider G (2007) Properties and architecture of drugs and natural products revisited. Curr Chem Biol 1: 115–127.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

