Characterization of the Ubiquitin-Conjugating Enzyme Gene Family in Rice and Evaluation of Expression Profiles under Abiotic Stresses and Hormone Treatments
- PMID: 25902049
- PMCID: PMC4406754
- DOI: 10.1371/journal.pone.0122621
Characterization of the Ubiquitin-Conjugating Enzyme Gene Family in Rice and Evaluation of Expression Profiles under Abiotic Stresses and Hormone Treatments
Abstract
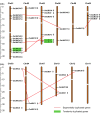
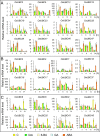
Ubiquitin-conjugating enzyme E2s (UBCs), which catalyze the transfer of ubiquitin to substrate or E3 ligases, are key enzymes in ubiquitination modifications of target proteins. However, little is known about the knowledge of UBC gene family in rice. In this study, a total of 39 UBC encoding genes, which all contained an UBC domain with a cysteine active site, were identified in the rice genome. These were classified into fifteen distinct subfamilies based upon their sequence similarity and phylogenetic relationships. A subset of 19 OsUBC genes exhibited chromosomal duplication; 4 and 15 OsUBC genes were tandemly and segmentally duplicated, respectively. Comprehensive analyses were performed to investigate the expression profiles of OsUBC genes in various stages of vegetative and reproductive development using data from EST, Microarrays, MPSS, and real-time PCR. Many OsUBC genes exhibited abundant and tissue-specific expression patterns. Moreover, 14 OsUBCs were found to be differentially expressed under treatments with drought, or salt stresses. The expression analysis after treatments with IAA, 6-BA, GA and ABA indicated that almost all OsUBC genes were responsive to at least two of the four hormones. Several genes were significantly down-regulated under all of the hormone treatments, and most of the genes reduced by 6-BA were also reduced by GA. This study will facilitate further studies of the OsUBC gene family and provide useful clues for functional validation of OsUBCs in rice.
Conflict of interest statement
Figures







References
-
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annual Review of Biochemistry 70: 503–533. - PubMed
-
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590. - PubMed
-
- Vierstra RD (1996) Proteolysis in plants: mechanisms and functions. Plant Mol Biol 32: 275–302. - PubMed
-
- Welchman R, Gordon C, Mayer J (2005) Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol 6:599–609. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

