Phosphoethanolamine Transferase LptA in Haemophilus ducreyi Modifies Lipid A and Contributes to Human Defensin Resistance In Vitro
- PMID: 25902140
- PMCID: PMC4406763
- DOI: 10.1371/journal.pone.0124373
Phosphoethanolamine Transferase LptA in Haemophilus ducreyi Modifies Lipid A and Contributes to Human Defensin Resistance In Vitro
Abstract
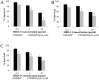
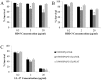
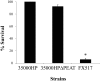
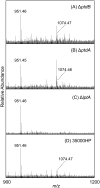
Haemophilus ducreyi resists the cytotoxic effects of human antimicrobial peptides (APs), including α-defensins, β-defensins, and the cathelicidin LL-37. Resistance to LL-37, mediated by the sensitive to antimicrobial peptide (Sap) transporter, is required for H. ducreyi virulence in humans. Cationic APs are attracted to the negatively charged bacterial cell surface. In other gram-negative bacteria, modification of lipopolysaccharide or lipooligosaccharide (LOS) by the addition of positively charged moieties, such as phosphoethanolamine (PEA), confers AP resistance by means of electrostatic repulsion. H. ducreyi LOS has PEA modifications at two sites, and we identified three genes (lptA, ptdA, and ptdB) in H. ducreyi with homology to a family of bacterial PEA transferases. We generated non-polar, unmarked mutants with deletions in one, two, or all three putative PEA transferase genes. The triple mutant was significantly more susceptible to both α- and β-defensins; complementation of all three genes restored parental levels of AP resistance. Deletion of all three PEA transferase genes also resulted in a significant increase in the negativity of the mutant cell surface. Mass spectrometric analysis revealed that LptA was required for PEA modification of lipid A; PtdA and PtdB did not affect PEA modification of LOS. In human inoculation experiments, the triple mutant was as virulent as its parent strain. While this is the first identified mechanism of resistance to α-defensins in H. ducreyi, our in vivo data suggest that resistance to cathelicidin LL-37 may be more important than defensin resistance to H. ducreyi pathogenesis.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

