Olfactomedin-1 Has a V-shaped Disulfide-linked Tetrameric Structure
- PMID: 25903135
- PMCID: PMC4463452
- DOI: 10.1074/jbc.M115.653485
Olfactomedin-1 Has a V-shaped Disulfide-linked Tetrameric Structure
Abstract
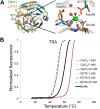
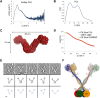
Olfactomedin-1 (Olfm1; also known as noelin and pancortin) is a member of the olfactomedin domain-containing superfamily and a highly expressed neuronal glycoprotein important for nervous system development. It binds a number of secreted proteins and cell surface-bound receptors to induce cell signaling processes. Using a combined approach of x-ray crystallography, solution scattering, analytical ultracentrifugation, and electron microscopy we determined that full-length Olfm1 forms disulfide-linked tetramers with a distinctive V-shaped architecture. The base of the "V" is formed by two disulfide-linked dimeric N-terminal domains. Each of the two V legs consists of a parallel dimeric disulfide-linked coiled coil with a C-terminal β-propeller dimer at the tips. This agrees with our crystal structure of a C-terminal coiled-coil segment and β-propeller combination (Olfm1(coil-Olf)) that reveals a disulfide-linked dimeric arrangement with the β-propeller top faces in an outward exposed orientation. Similar to its family member myocilin, Olfm1 is stabilized by calcium. The dimer-of-dimers architecture suggests a role for Olfm1 in clustering receptors to regulate signaling and sheds light on the conformation of several other olfactomedin domain family members.
Keywords: analytical ultracentrifugation; cell signaling; coiled coil; development; disulfide; electron tomography; neurobiology; olfactomedin-1 (Olfm1); small angle x-ray scattering (SAXS); x-ray crystallography.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

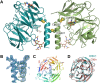


References
-
- Snyder D. A., Rivers A. M., Yokoe H., Menco B. P., Anholt R. R. (1991) Olfactomedin: purification, characterization, and localization of a novel olfactory glycoprotein. Biochemistry. 30, 9143–9153 - PubMed
-
- Moreno T. A., Bronner-Fraser M. (2001) The secreted glycoprotein Noelin-1 promotes neurogenesis in Xenopus. Dev. Biol. 240, 340–360 - PubMed
-
- Moreno T. A., Bronner-Fraser M. (2005) Noelins modulate the timing of neuronal differentiation during development. Dev. Biol. 288, 434–447 - PubMed
-
- Barembaum M., Moreno T. A., LaBonne C., Sechrist J., Bronner-Fraser M. (2000) Noelin-1 is a secreted glycoprotein involved in generation of the neural crest. Nat. Cell Biol. 2, 219–225 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

