Deciphering poxvirus gene expression by RNA sequencing and ribosome profiling
- PMID: 25903347
- PMCID: PMC4468498
- DOI: 10.1128/JVI.00528-15
Deciphering poxvirus gene expression by RNA sequencing and ribosome profiling
Abstract
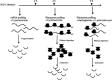
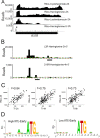
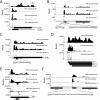
The more than 200 closely spaced annotated open reading frames, extensive transcriptional read-through, and numerous unpredicted RNA start sites have made the analysis of vaccinia virus gene expression challenging. Genome-wide ribosome profiling provided an unprecedented assessment of poxvirus gene expression. By 4 h after infection, approximately 80% of the ribosome-associated mRNA was viral. Ribosome-associated mRNAs were detected for most annotated early genes at 2 h and for most intermediate and late genes at 4 and 8 h. Cluster analysis identified a subset of early mRNAs that continued to be translated at the later times. At 2 h, there was excellent correlation between the abundance of individual mRNAs and the numbers of associated ribosomes, indicating that expression was primarily transcriptionally regulated. However, extensive transcriptional read-through invalidated similar correlations at later times. The mRNAs with the highest density of ribosomes had host response, DNA replication, and transcription roles at early times and were virion components at late times. Translation inhibitors were used to map initiation sites at single-nucleotide resolution at the start of most annotated open reading frames although in some cases a downstream methionine was used instead. Additional putative translational initiation sites with AUG or alternative codons occurred mostly within open reading frames, and fewer occurred in untranslated leader sequences, antisense strands, and intergenic regions. However, most open reading frames associated with these additional translation initiation sites were short, raising questions regarding their biological roles. The data were used to construct a high-resolution genome-wide map of the vaccinia virus translatome.
Importance: This report contains the first genome-wide, high-resolution analysis of poxvirus gene expression at both transcriptional and translational levels. The study was made possible by recent methodological advances allowing examination of the translated regions of mRNAs including start sites at single-nucleotide resolution. Vaccinia virus ribosome-associated mRNA sequences were detected for most annotated early genes at 2 h and for most intermediate and late genes at 4 and 8 h after infection. The ribosome profiling approach was particularly valuable for poxviruses because of the close spacing of approximately 200 open reading frames and extensive transcriptional read-through resulting in overlapping mRNAs. The expression of intermediate and late genes, in particular, was visualized with unprecedented clarity and quantitation. We also identified novel putative translation initiation sites that were mostly associated with short protein coding sequences. The results provide a framework for further studies of poxvirus gene expression.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures





References
-
- Moss B. 2013. Poxviridae, p 2129–2159. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Rancaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Damon I. 2013. Poxviruses, p 2160–2184. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Rancaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Martin SA, Paoletti E, Moss B. 1975. Purification of mRNA guanylyltransferase and mRNA (guanine 7-)methyltransferase from vaccinia virus. J Biol Chem 250:9322–9329. - PubMed
-
- Kates J, Beeson J. 1970. Ribonucleic acid synthesis in vaccinia virus. II. Synthesis of polyriboadenylic acid. J Mol Biol 50:19–23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

