Comparative genomic analysis reveals occurrence of genetic recombination in virulent Cryptosporidium hominis subtypes and telomeric gene duplications in Cryptosporidium parvum
- PMID: 25903370
- PMCID: PMC4407392
- DOI: 10.1186/s12864-015-1517-1
Comparative genomic analysis reveals occurrence of genetic recombination in virulent Cryptosporidium hominis subtypes and telomeric gene duplications in Cryptosporidium parvum
Abstract
Background: Cryptosporidium hominis is a dominant species for human cryptosporidiosis. Within the species, IbA10G2 is the most virulent subtype responsible for all C. hominis-associated outbreaks in Europe and Australia, and is a dominant outbreak subtype in the United States. In recent yearsIaA28R4 is becoming a major new subtype in the United States. In this study, we sequenced the genomes of two field specimens from each of the two subtypes and conducted a comparative genomic analysis of the obtained sequences with those from the only fully sequenced Cryptosporidium parvum genome.
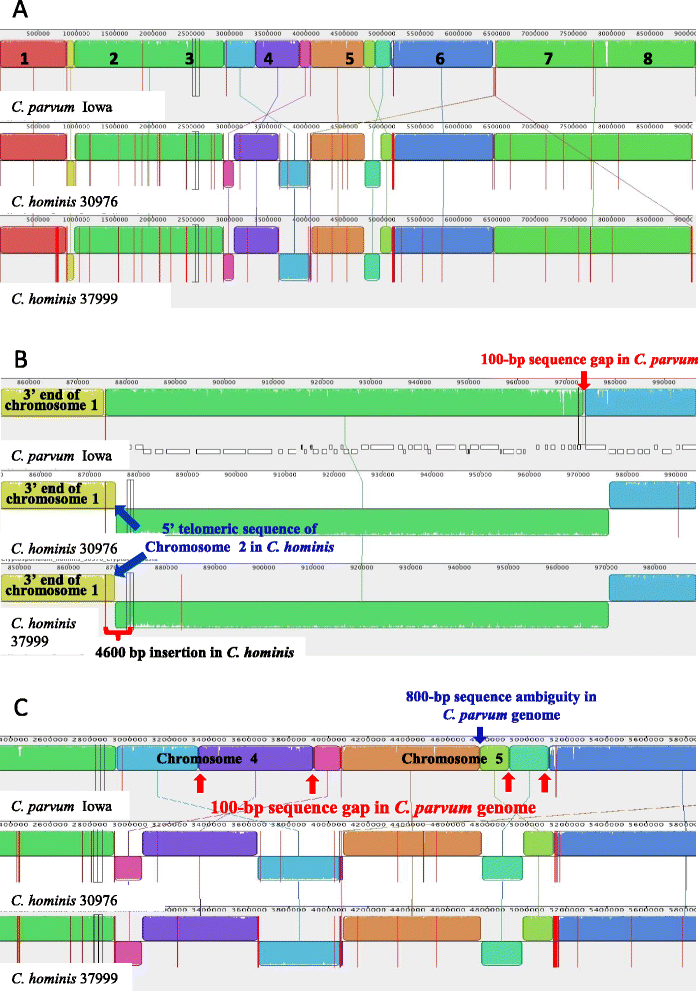
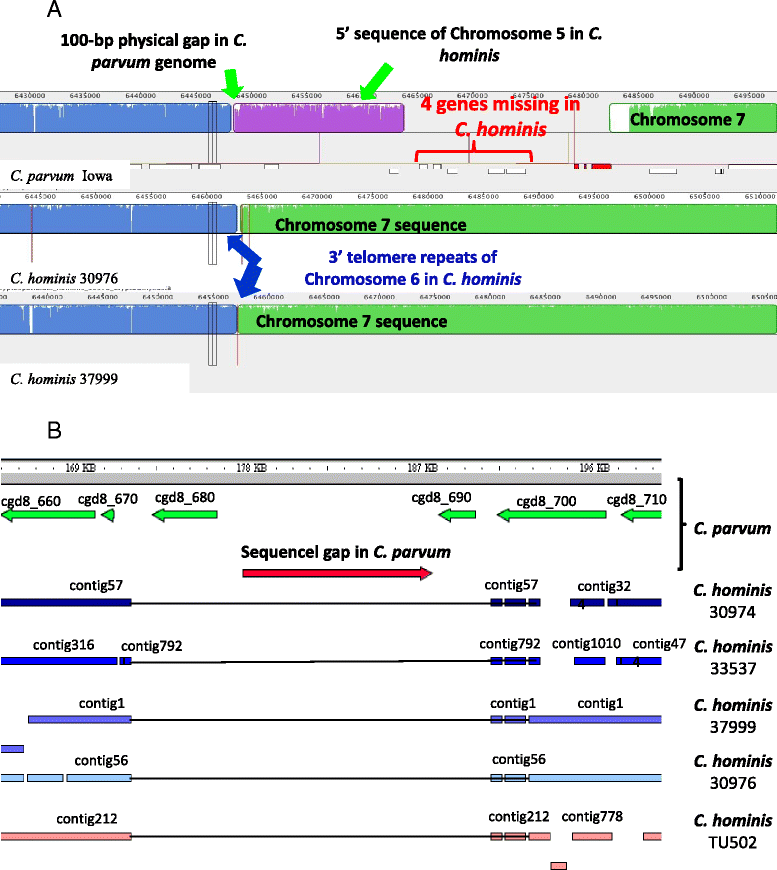
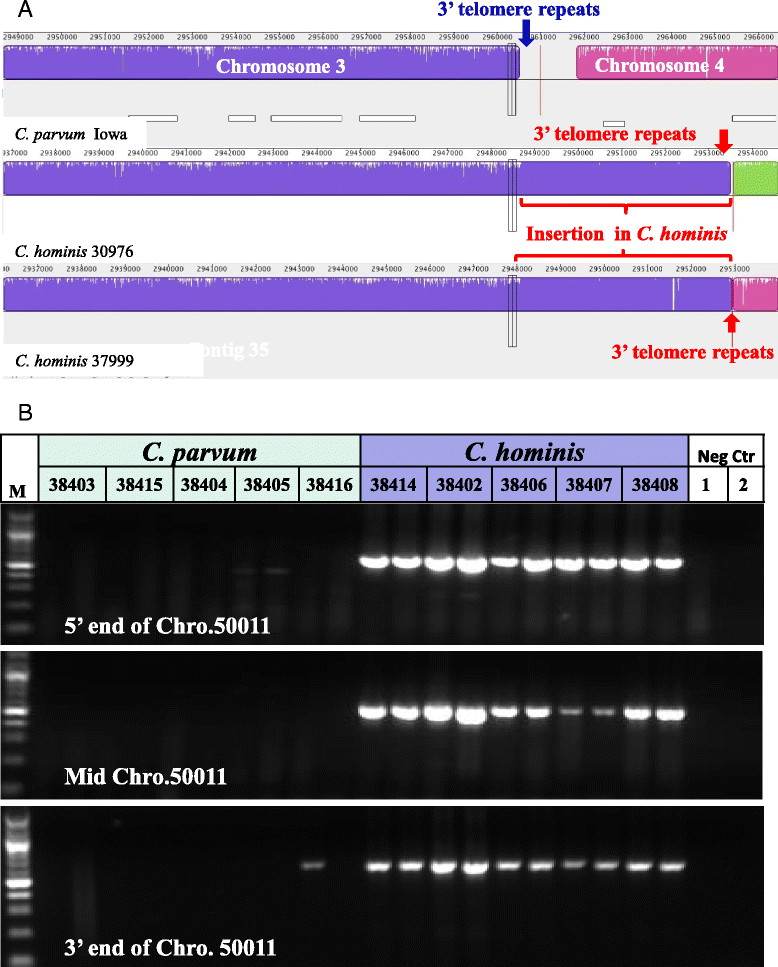
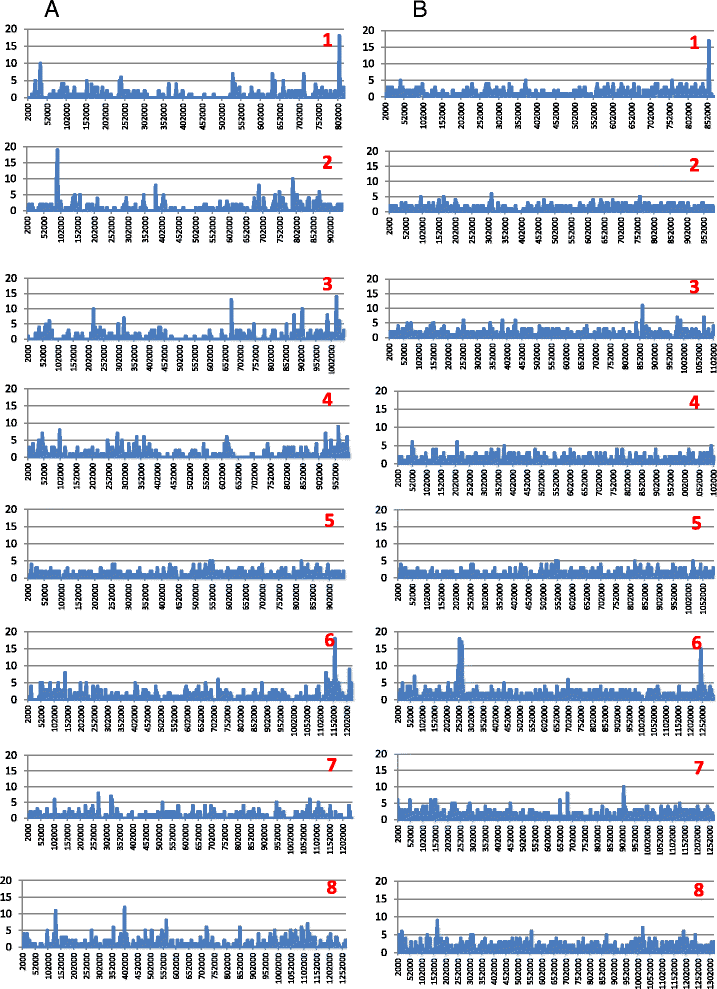
Results: Altogether, 8.59-9.05 Mb of Cryptosporidium sequences in 45-767 assembled contigs were obtained from the four specimens, representing 94.36-99.47% coverage of the expected genome. These genomes had complete synteny in gene organization and 96.86-97.0% and 99.72-99.83% nucleotide sequence similarities to the published genomes of C. parvum and C. hominis, respectively. Several major insertions and deletions were seen between C. hominis and C. parvum genomes, involving mostly members of multicopy gene families near telomeres. The four C. hominis genomes were highly similar to each other and divergent from the reference IaA25R3 genome in some highly polymorphic regions. Major sequence differences among the four specimens sequenced in this study were in the 5' and 3' ends of chromosome 6 and the gp60 region, largely the result of genetic recombination.
Conclusions: The sequence similarity among specimens of the two dominant outbreak subtypes and genetic recombination in chromosome 6, especially around the putative virulence determinant gp60 region, suggest that genetic recombination plays a potential role in the emergence of hyper-transmissible C. hominis subtypes. The high sequence conservation between C. parvum and C. hominis genomes and significant differences in copy numbers of MEDLE family secreted proteins and insulinase-like proteases indicate that telomeric gene duplications could potentially contribute to host expansion in C. parvum.
Figures
References
-
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case–control study. Lancet. 2013;382(9888):209–22. doi: 10.1016/S0140-6736(13)60844-2. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

