Measuring Food Intake and Nutrient Absorption in Caenorhabditis elegans
- PMID: 25903497
- PMCID: PMC4492371
- DOI: 10.1534/genetics.115.175851
Measuring Food Intake and Nutrient Absorption in Caenorhabditis elegans
Abstract
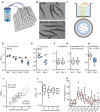
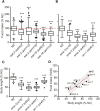
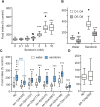
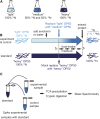
Caenorhabditis elegans has emerged as a powerful model to study the genetics of feeding, food-related behaviors, and metabolism. Despite the many advantages of C. elegans as a model organism, direct measurement of its bacterial food intake remains challenging. Here, we describe two complementary methods that measure the food intake of C. elegans. The first method is a microtiter plate-based bacterial clearing assay that measures food intake by quantifying the change in the optical density of bacteria over time. The second method, termed pulse feeding, measures the absorption of food by tracking de novo protein synthesis using a novel metabolic pulse-labeling strategy. Using the bacterial clearance assay, we compare the bacterial food intake of various C. elegans strains and show that long-lived eat mutants eat substantially more than previous estimates. To demonstrate the applicability of the pulse-feeding assay, we compare the assimilation of food for two C. elegans strains in response to serotonin. We show that serotonin-increased feeding leads to increased protein synthesis in a SER-7-dependent manner, including proteins known to promote aging. Protein content in the food has recently emerged as critical factor in determining how food composition affects aging and health. The pulse-feeding assay, by measuring de novo protein synthesis, represents an ideal method to unequivocally establish how the composition of food dictates protein synthesis. In combination, these two assays provide new and powerful tools for C. elegans research to investigate feeding and how food intake affects the proteome and thus the physiology and health of an organism.
Keywords: aging; de novo protein synthesis; feeding; metabolism; nutrition.
Copyright © 2015 by the Genetics Society of America.
Figures
References
-
- Ashrafi K., 2006. Mapping out starvation responses. Cell Metab. 3: 235–236. - PubMed
-
- Ashrafi K., Chang F. Y., Watts J. L., Fraser A. G., Kamath R. S., et al. , 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421: 268–272. - PubMed
-
- Beaulieu J. M., Sotnikova T. D., Marion S., Lefkowitz R. J., Gainetdinov R. R., et al. , 2005. An Akt/beta-arrestin 2/PP2A signaling complex mediates dopaminergic neurotransmission and behavior. Cell 122: 261–273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

