KaBOB: ontology-based semantic integration of biomedical databases
- PMID: 25903923
- PMCID: PMC4448321
- DOI: 10.1186/s12859-015-0559-3
KaBOB: ontology-based semantic integration of biomedical databases
Abstract
Background: The ability to query many independent biological databases using a common ontology-based semantic model would facilitate deeper integration and more effective utilization of these diverse and rapidly growing resources. Despite ongoing work moving toward shared data formats and linked identifiers, significant problems persist in semantic data integration in order to establish shared identity and shared meaning across heterogeneous biomedical data sources.
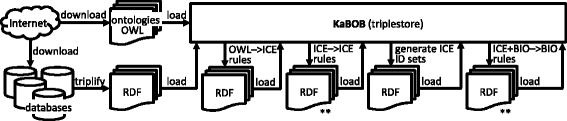
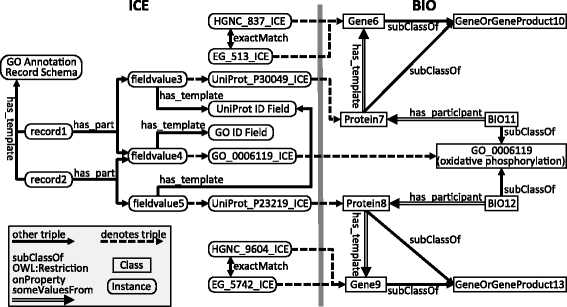
Results: We present five processes for semantic data integration that, when applied collectively, solve seven key problems. These processes include making explicit the differences between biomedical concepts and database records, aggregating sets of identifiers denoting the same biomedical concepts across data sources, and using declaratively represented forward-chaining rules to take information that is variably represented in source databases and integrating it into a consistent biomedical representation. We demonstrate these processes and solutions by presenting KaBOB (the Knowledge Base Of Biomedicine), a knowledge base of semantically integrated data from 18 prominent biomedical databases using common representations grounded in Open Biomedical Ontologies. An instance of KaBOB with data about humans and seven major model organisms can be built using on the order of 500 million RDF triples. All source code for building KaBOB is available under an open-source license.
Conclusions: KaBOB is an integrated knowledge base of biomedical data representationally based in prominent, actively maintained Open Biomedical Ontologies, thus enabling queries of the underlying data in terms of biomedical concepts (e.g., genes and gene products, interactions and processes) rather than features of source-specific data schemas or file formats. KaBOB resolves many of the issues that routinely plague biomedical researchers intending to work with data from multiple data sources and provides a platform for ongoing data integration and development and for formal reasoning over a wealth of integrated biomedical data.
Figures
Similar articles
-
Using the Semantic Web for Rapid Integration of WikiPathways with Other Biological Online Data Resources.PLoS Comput Biol. 2016 Jun 23;12(6):e1004989. doi: 10.1371/journal.pcbi.1004989. eCollection 2016 Jun. PLoS Comput Biol. 2016. PMID: 27336457 Free PMC article.
-
AlzPharm: integration of neurodegeneration data using RDF.BMC Bioinformatics. 2007 May 9;8 Suppl 3(Suppl 3):S4. doi: 10.1186/1471-2105-8-S3-S4. BMC Bioinformatics. 2007. PMID: 17493287 Free PMC article.
-
Semantic web for integrated network analysis in biomedicine.Brief Bioinform. 2009 Mar;10(2):177-92. doi: 10.1093/bib/bbp002. Brief Bioinform. 2009. PMID: 19304873
-
Advancing translational research with the Semantic Web.BMC Bioinformatics. 2007 May 9;8 Suppl 3(Suppl 3):S2. doi: 10.1186/1471-2105-8-S3-S2. BMC Bioinformatics. 2007. PMID: 17493285 Free PMC article. Review.
-
Techniques for optimization of queries on integrated biological resources.J Bioinform Comput Biol. 2004 Jun;2(2):375-411. doi: 10.1142/s0219720004000648. J Bioinform Comput Biol. 2004. PMID: 15297988 Review.
Cited by
-
Establishing a consensus for the hallmarks of cancer based on gene ontology and pathway annotations.BMC Bioinformatics. 2021 Apr 6;22(1):178. doi: 10.1186/s12859-021-04105-8. BMC Bioinformatics. 2021. PMID: 33823788 Free PMC article.
-
CROssBAR: comprehensive resource of biomedical relations with knowledge graph representations.Nucleic Acids Res. 2021 Sep 20;49(16):e96. doi: 10.1093/nar/gkab543. Nucleic Acids Res. 2021. PMID: 34181736 Free PMC article.
-
SMASH: A Data-driven Informatics Method to Assist Experts in Characterizing Semantic Heterogeneity among Data Elements.AMIA Annu Symp Proc. 2017 Feb 10;2016:1717-1726. eCollection 2016. AMIA Annu Symp Proc. 2017. PMID: 28269930 Free PMC article.
-
The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species.Nucleic Acids Res. 2017 Jan 4;45(D1):D712-D722. doi: 10.1093/nar/gkw1128. Epub 2016 Nov 29. Nucleic Acids Res. 2017. PMID: 27899636 Free PMC article.
-
Therapeutic futility and phenotypic heterogeneity in heart failure with preserved ejection fraction: what is the role of bionic learning?Eur J Heart Fail. 2020 Jan;22(1):159-161. doi: 10.1002/ejhf.1658. Epub 2019 Nov 20. Eur J Heart Fail. 2020. PMID: 31749260 Free PMC article. No abstract available.
References
-
- Jain P, Hitzler P, Yeh PZ, Verma K, Sheth AP, Linked Data Is Merely More Data. In: Dan Brickley, Vinay K. Chaudhri, Harry Halpin, and Deborah McGuinness: Linked Data Meets Artificial Intelligence. Technical Report SS-10-07, AAAI Press, Menlo Park, California, 2010, pp.82-86. ISBN 978-1-57735-461-1
-
- Hitzler, P. Towards reasoning pragmatics. In: Janowicz, K., Raubal, M., Levashkin, S. (eds.) GeoSpatial Semantics, Third International Conference, GeoS 2009, Mexico City, Mexico, December 3–4, 2009. Proceedings. pp. 9–25. Lecture Notes in Computer Science, Springer (2009)
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

