Isolation, abundance and phylogenetic affiliation of endophytic actinomycetes associated with medicinal plants and screening for their in vitro antimicrobial biosynthetic potential
- PMID: 25904906
- PMCID: PMC4388002
- DOI: 10.3389/fmicb.2015.00273
Isolation, abundance and phylogenetic affiliation of endophytic actinomycetes associated with medicinal plants and screening for their in vitro antimicrobial biosynthetic potential
Abstract
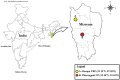
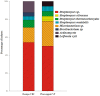
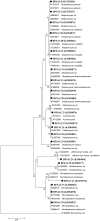
Microorganisms associated with medicinal plants are of interest as the producers of important bioactive compounds. To date, the diversity of culturable endophytic actinomycetes associated with medicinal plants is in its initial phase of exploration. In this study, 42 endophytic actinomycetes were isolated from different organs of seven selected medicinal plants. The highest number of isolates (n = 22, 52.3%) of actinomycetes was isolated from roots, followed by stems (n = 9, 21.4%), leaves (n = 6, 14.2%), flowers (n = 3, 7.1%), and petioles (n = 2, 4.7%). The genus Streptomyces was the most dominant among the isolates (66.6%) in both the locations (Dampa TRF and Phawngpuii NP, Mizoram, India). From a total of 42 isolates, 22 isolates were selected for further studies based on their ability to inhibit one of the tested human bacterial or fungal pathogen. Selected isolates were identified based on 16S rRNA gene analysis and subsequently the isolates were grouped to four different genera; Streptomyces, Brevibacterium, Microbacterium, and Leifsonia. Antibiotic sensitivity assay was performed to understand the responsible antimicrobials present in the isolates showing the antimicrobial activities and revealed that the isolates were mostly resistant to penicillin G and ampicillin. Further, antimicrobial properties and antibiotic sensitivity assay in combination with the results of amplification of biosynthetic genes polyketide synthase (PKS-I) and non-ribosomal peptide synthetase (NRPS) showed that the endophytic actinomycetes associated with the selected medicinal plants have broad-spectrum antimicrobial activity. This is the first report of the isolation of Brevibacterium sp., Microbacterium sp., and Leifsonia xyli from endophytic environments of medicinal plants, Mirabilis jalapa and Clerodendrum colebrookianum. Our results emphasize that endophytic actinomycetes associated with medicinal plants are an unexplored resource for the discovery of biologically active compounds.
Keywords: 16S rRNA gene; antibiotic sensitivity; endophytic actinomycetes; non-ribosomal peptide synthetase (NRPS); polyketide synthase (PKS-I).
Figures

Similar articles
-
Endophytic actinomycetes associated with Cinnamomum cassia Presl in Hoa Binh province, Vietnam: Distribution, antimicrobial activity and, genetic features.J Gen Appl Microbiol. 2020 Apr 13;66(1):24-31. doi: 10.2323/jgam.2019.04.004. Epub 2019 Aug 2. J Gen Appl Microbiol. 2020. PMID: 31378748
-
Antimicrobial biosynthetic potential and genetic diversity of endophytic actinomycetes associated with medicinal plants.FEMS Microbiol Lett. 2015 Oct;362(19):fnv158. doi: 10.1093/femsle/fnv158. Epub 2015 Sep 6. FEMS Microbiol Lett. 2015. PMID: 26347302
-
Endophytic Actinomycetes from Tea Plants (Camellia sinensis): Isolation, Abundance, Antimicrobial, and Plant-Growth-Promoting Activities.Biomed Res Int. 2018 Nov 1;2018:1470305. doi: 10.1155/2018/1470305. eCollection 2018. Biomed Res Int. 2018. PMID: 30519568 Free PMC article.
-
Antimicrobial Action Mechanisms of Natural Compounds Isolated from Endophytic Microorganisms.Antibiotics (Basel). 2024 Mar 18;13(3):271. doi: 10.3390/antibiotics13030271. Antibiotics (Basel). 2024. PMID: 38534706 Free PMC article. Review.
-
Endophytic bacteria: a new source of bioactive compounds.3 Biotech. 2017 Oct;7(5):315. doi: 10.1007/s13205-017-0942-z. Epub 2017 Sep 14. 3 Biotech. 2017. PMID: 28955612 Free PMC article. Review.
Cited by
-
P-Solubilizing Streptomyces roseocinereus MS1B15 With Multiple Plant Growth-Promoting Traits Enhance Barley Development and Regulate Rhizosphere Microbial Population.Front Plant Sci. 2020 Aug 7;11:1137. doi: 10.3389/fpls.2020.01137. eCollection 2020. Front Plant Sci. 2020. PMID: 32849698 Free PMC article.
-
Implication of PKS type I gene and chromatographic strategy for the biodiscovery of antimicrobial polyketide metabolites from endosymbiotic Nocardiopsis prasina CLA68.Naturwissenschaften. 2016 Jun;103(5-6):45. doi: 10.1007/s00114-016-1370-3. Epub 2016 May 6. Naturwissenschaften. 2016. PMID: 27154505
-
Continuing hunt for endophytic actinomycetes as a source of novel biologically active metabolites.World J Microbiol Biotechnol. 2015 Dec;31(12):1863-75. doi: 10.1007/s11274-015-1950-y. Epub 2015 Sep 26. World J Microbiol Biotechnol. 2015. PMID: 26410426
-
Identification, Biocontrol and Plant Growth Promotion Potential of Endophytic Streptomyces sp. a13.Curr Microbiol. 2025 Jan 3;82(2):64. doi: 10.1007/s00284-024-04009-9. Curr Microbiol. 2025. PMID: 39751911
-
Bioprospecting of endophytic actinobacterium associated with Aloe ferox mill for antibacterial activity.BMC Complement Med Ther. 2022 Oct 3;22(1):258. doi: 10.1186/s12906-022-03733-8. BMC Complement Med Ther. 2022. PMID: 36192707 Free PMC article.
References
-
- Akintobi O. A., Agunbiade S. O., Okonko I. O., Ojo O. V. (2011). Antimicrobial evaluation and phytochemical analysis of leaf extracts of Mirabilis jalapa against some human pathogenic bacteria. Nat. Sci. 9, 45–53.
-
- Bergey D. H., Holt J. G. (2000). Bergey's Manual of Determinative Bacteriology, 9th Edn Philadelphia, PA: Lippincott Williams and Wilkins.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

