Strategies for B-cell receptor repertoire analysis in primary immunodeficiencies: from severe combined immunodeficiency to common variable immunodeficiency
- PMID: 25904919
- PMCID: PMC4389565
- DOI: 10.3389/fimmu.2015.00157
Strategies for B-cell receptor repertoire analysis in primary immunodeficiencies: from severe combined immunodeficiency to common variable immunodeficiency
Abstract
The antigen receptor repertoires of B- and T-cells form the basis of the adaptive immune response. The repertoires should be sufficiently diverse to recognize all possible pathogens. However, careful selection is needed to prevent responses to self or harmless antigens. Limited antigen receptor repertoire diversity leads to immunodeficiency, whereas unselected or misdirected repertoires can result in autoimmunity. The antigen receptor repertoire harbors information about abnormalities in many immunological disorders. Recent developments in next generation sequencing allow the analysis of the antigen receptor repertoire in much greater detail than ever before. Analyzing the antigen receptor repertoire in patients with mutations in genes responsible for the generation of the antigen receptor repertoire will give new insights into repertoire formation and selection. In this perspective, we describe strategies and considerations for analysis of the naive and antigen-selected B-cell repertoires in primary immunodeficiency patients with a focus on severe combined immunodeficiency and common variable immunodeficiency.
Keywords: CVID; V(D)J recombination; immunodeficiency; next generation sequencing; repertoire.
Figures
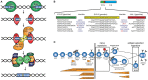
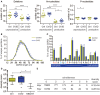
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

