XomAnnotate: Analysis of Heterogeneous and Complex Exome- A Step towards Translational Medicine
- PMID: 25905921
- PMCID: PMC4408095
- DOI: 10.1371/journal.pone.0123569
XomAnnotate: Analysis of Heterogeneous and Complex Exome- A Step towards Translational Medicine
Abstract
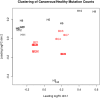
In translational cancer medicine, implicated pathways and the relevant master genes are of focus. Exome's specificity, processing-time, and cost advantage makes it a compelling tool for this purpose. However, analysis of exome lacks reliable combinatory analysis tools and techniques. In this paper we present XomAnnotate--a meta- and functional-analysis software for exome. We compared UnifiedGenotyper, Freebayes, Delly, and Lumpy algorithms that were designed for whole-genome and combined their strengths in XomAnnotate for exome data through meta-analysis to identify comprehensive mutation profile (SNPs/SNVs, short inserts/deletes, and SVs) of patients. The mutation profile is annotated followed by functional analysis through pathway enrichment and network analysis to identify most critical genes and pathways implicated in the disease genesis. The efficacy of the software is verified through MDS and clustering and tested with available 11 familial non-BRCA1/BRCA2 breast cancer exome data. The results showed that the most significantly affected pathways across all samples are cell communication and antigen processing and presentation. ESCO1, HYAL1, RAF1 and PRKCA emerged as the key genes. Network analysis further showed the purine and propanotate metabolism pathways along with RAF1 and PRKCA genes to be master regulators in these patients. Therefore, XomAnnotate is able to use exome data to identify entire mutation landscape, pathways, and the master genes accurately with wide concordance from earlier microarray and whole-genome studies--making it a suitable biomedical software for using exome in next-generation translational medicine.
Conflict of interest statement
Figures


Similar articles
-
Whole exome sequencing of rare aggressive breast cancer histologies.Breast Cancer Res Treat. 2016 Feb;156(1):21-32. doi: 10.1007/s10549-016-3718-y. Epub 2016 Feb 23. Breast Cancer Res Treat. 2016. PMID: 26907767
-
Whole-exome sequencing reveals recurrent somatic mutation networks in cancer.Cancer Lett. 2013 Nov 1;340(2):270-6. doi: 10.1016/j.canlet.2012.11.002. Epub 2012 Nov 12. Cancer Lett. 2013. PMID: 23153794 Review.
-
Exome sequencing reveals frequent deleterious germline variants in cancer susceptibility genes in women with invasive breast cancer undergoing neoadjuvant chemotherapy.Breast Cancer Res Treat. 2015 Sep;153(2):435-43. doi: 10.1007/s10549-015-3545-6. Epub 2015 Aug 22. Breast Cancer Res Treat. 2015. PMID: 26296701 Free PMC article.
-
Translating exome sequencing from research to clinical diagnostics.Clin Chem Lab Med. 2011 Dec 23;50(7):1161-8. doi: 10.1515/cclm-2011-0841. Clin Chem Lab Med. 2011. PMID: 22850020 Review.
-
Characterization of familial breast cancer in Saudi Arabia.BMC Genomics. 2015;16 Suppl 1(Suppl 1):S3. doi: 10.1186/1471-2164-16-S1-S3. Epub 2015 Jan 15. BMC Genomics. 2015. PMID: 25923920 Free PMC article.
Cited by
-
Tracking Cancer Genetic Evolution using OncoTrack.Sci Rep. 2016 Jul 14;6:29647. doi: 10.1038/srep29647. Sci Rep. 2016. PMID: 27412732 Free PMC article.
-
A Survey of Computational Tools to Analyze and Interpret Whole Exome Sequencing Data.Int J Genomics. 2016;2016:7983236. doi: 10.1155/2016/7983236. Epub 2016 Dec 14. Int J Genomics. 2016. PMID: 28070503 Free PMC article. Review.
-
Molecular characterization of organoids derived from pancreatic intraductal papillary mucinous neoplasms.J Pathol. 2020 Nov;252(3):252-262. doi: 10.1002/path.5515. Epub 2020 Sep 19. J Pathol. 2020. PMID: 32696980 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

