Protein degradation pathways regulate the functions of helicases in the DNA damage response and maintenance of genomic stability
- PMID: 25906194
- PMCID: PMC4496686
- DOI: 10.3390/biom5020590
Protein degradation pathways regulate the functions of helicases in the DNA damage response and maintenance of genomic stability
Abstract
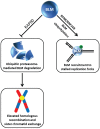
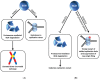
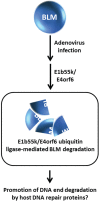
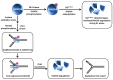
Degradation of helicases or helicase-like proteins, often mediated by ubiquitin-proteasomal pathways, plays important regulatory roles in cellular mechanisms that respond to DNA damage or replication stress. The Bloom's syndrome helicase (BLM) provides an example of how helicase degradation pathways, regulated by post-translational modifications and protein interactions with components of the Fanconi Anemia (FA) interstrand cross-link (ICL) repair pathway, influence cell cycle checkpoints, DNA repair, and replication restart. The FANCM DNA translocase can be targeted by checkpoint kinases that exert dramatic effects on FANCM stability and chromosomal integrity. Other work provides evidence that degradation of the F-box DNA helicase (FBH1) helps to balance translesion synthesis (TLS) and homologous recombination (HR) repair at blocked replication forks. Degradation of the helicase-like transcription factor (HLTF), a DNA translocase and ubiquitylating enzyme, influences the choice of post replication repair (PRR) pathway. Stability of the Werner syndrome helicase-nuclease (WRN) involved in the replication stress response is regulated by its acetylation. Turning to transcription, stability of the Cockayne Syndrome Group B DNA translocase (CSB) implicated in transcription-coupled repair (TCR) is regulated by a CSA ubiquitin ligase complex enabling recovery of RNA synthesis. Collectively, these studies demonstrate that helicases can be targeted for degradation to maintain genome homeostasis.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

