Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes
- PMID: 25907113
- PMCID: PMC4551309
- DOI: 10.1093/femsre/fuv015
Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes
Abstract
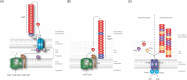
Biofilms are communities of microbial cells that underpin diverse processes including sewage bioremediation, plant growth promotion, chronic infections and industrial biofouling. The cells resident in the biofilm are encased within a self-produced exopolymeric matrix that commonly comprises lipids, proteins that frequently exhibit amyloid-like properties, eDNA and exopolysaccharides. This matrix fulfils a variety of functions for the community, from providing structural rigidity and protection from the external environment to controlling gene regulation and nutrient adsorption. Critical to the development of novel strategies to control biofilm infections, or the capability to capitalize on the power of biofilm formation for industrial and biotechnological uses, is an in-depth knowledge of the biofilm matrix. This is with respect to the structure of the individual components, the nature of the interactions between the molecules and the three-dimensional spatial organization. We highlight recent advances in the understanding of the structural and functional role that carbohydrates and proteins play within the biofilm matrix to provide three-dimensional architectural integrity and functionality to the biofilm community. We highlight, where relevant, experimental techniques that are allowing the boundaries of our understanding of the biofilm matrix to be extended using Escherichia coli, Staphylococcus aureus, Vibrio cholerae, and Bacillus subtilis as exemplars.
Keywords: Bacillus subtilis; Escherichia coli; Staphylococcus aureus; Vibrio cholerae; amyloid fibres; biofilm matrix assembly; biophysics; hydrophobin.
© FEMS 2015.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

