Rapid and comprehensive 'shotgun' lipidome profiling of colorectal cancer cell derived exosomes
- PMID: 25907253
- PMCID: PMC4615275
- DOI: 10.1016/j.ymeth.2015.04.014
Rapid and comprehensive 'shotgun' lipidome profiling of colorectal cancer cell derived exosomes
Abstract
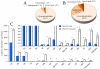
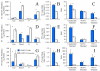
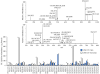
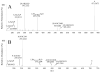
There is an increasing recognition of the role that cancer cell derived exosomes play in intercellular signaling upon fusion or uptake with a target cell, including immune system evasion, tumor growth and metastasis. To date, however, although exosomal membrane and cargo lipids are expected to play a pivotal role in exosome biogenesis and secretion, as well as in fusion or uptake and target cell functional response, the detailed characterization of cancer cell derived exosome lipids across a range of different cancers has not yet been broadly explored. Here, a simple and straightforward lipidome analysis strategy consisting of optimized sample extraction and novel sample derivatization techniques, coupled with high-resolution 'shotgun' mass spectrometry and 'targeted' tandem mass spectrometry methods, is demonstrated for the rapid identification of >520 individual lipids in 36 lipid classes and sub classes from exosomes secreted by the colorectal cancer cell line, LIM1215. Relative quantification and comparison of exosome versus cellular lipid profiles reveals significant enrichment of certain lipid classes, as well as substantial lipid subclass remodeling and changes in abundance of individual lipids, including sphingolipids, sterol lipids, glycerolipids and glycerophospholipids, and particularly plasmalogen- and alkyl ether-containing glycerophospholipids. This analysis strategy therefore provides a platform for comprehensive lipidome profiling across a wide range of cancer cell or tissue derived exosomes, that will facilitate subsequent functional studies aimed at elucidating the role of specific cellular or exosome lipids in the onset and progression of colorectal cancer, or to identify specific lipid(s) that could serve as effective diagnostic or prognostic disease biomarkers.
Keywords: Colorectal cancer; Exosome; Lipidome; Mass spectrometry.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

