A depauperate immune repertoire precedes evolution of sociality in bees
- PMID: 25908406
- PMCID: PMC4408586
- DOI: 10.1186/s13059-015-0628-y
A depauperate immune repertoire precedes evolution of sociality in bees
Abstract
Background: Sociality has many rewards, but can also be dangerous, as high population density and low genetic diversity, common in social insects, is ideal for parasite transmission. Despite this risk, honeybees and other sequenced social insects have far fewer canonical immune genes relative to solitary insects. Social protection from infection, including behavioral responses, may explain this depauperate immune repertoire. Here, based on full genome sequences, we describe the immune repertoire of two ecologically and commercially important bumblebee species that diverged approximately 18 million years ago, the North American Bombus impatiens and European Bombus terrestris.
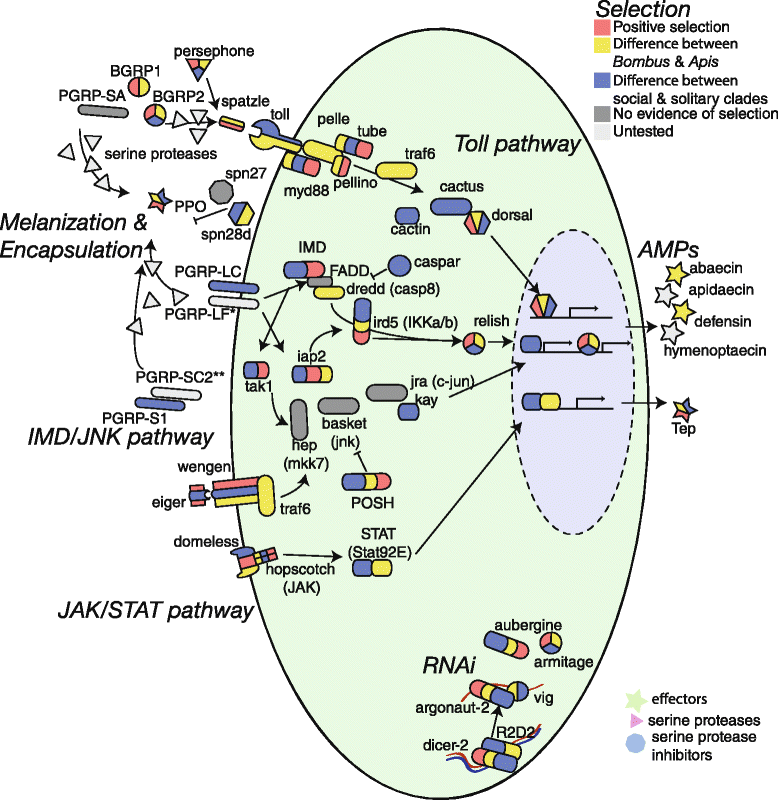
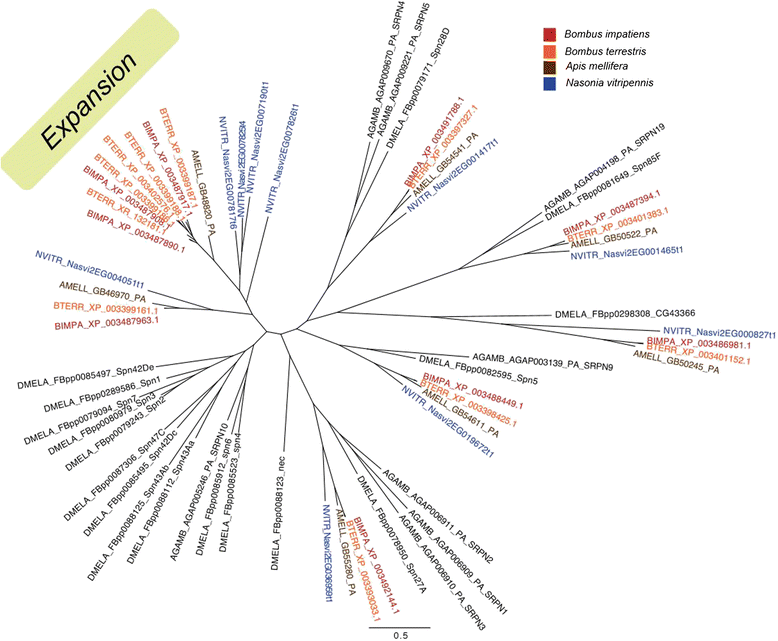
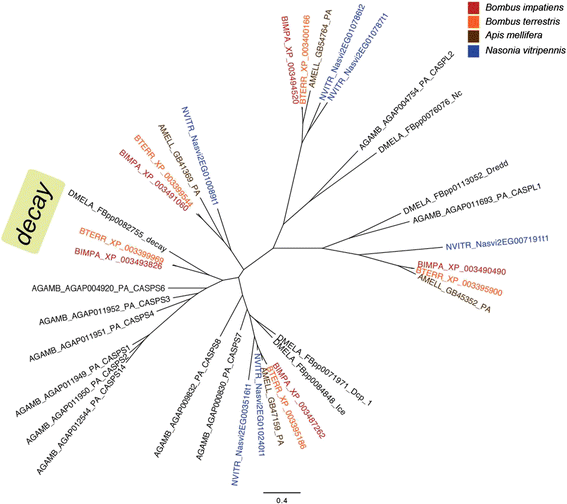
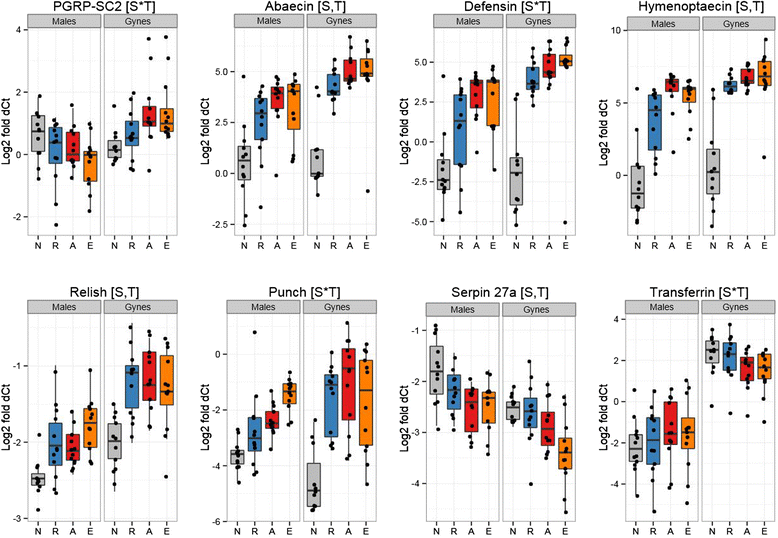
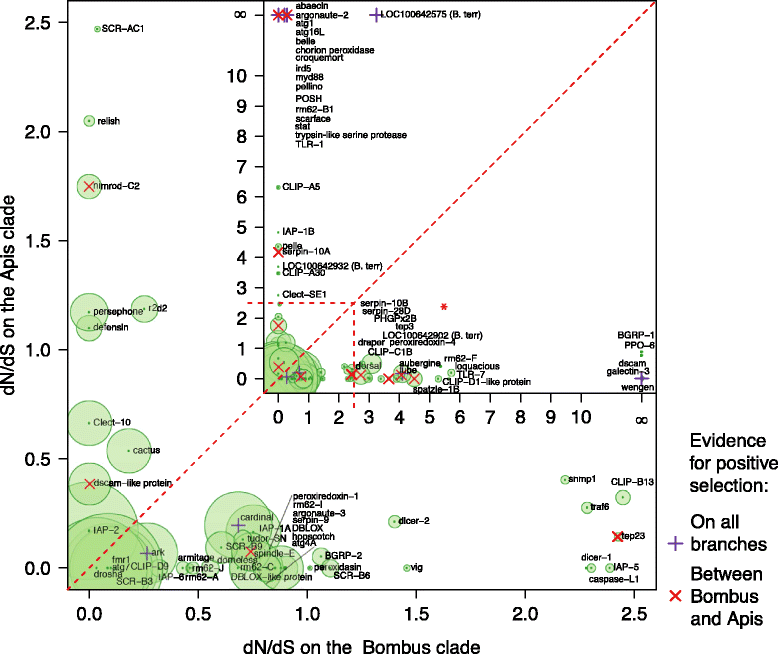
Results: We find that the immune systems of these bumblebees, two species of honeybee, and a solitary leafcutting bee, are strikingly similar. Transcriptional assays confirm the expression of many of these genes in an immunological context and more strongly in young queens than males, affirming Bateman's principle of greater investment in female immunity. We find evidence of positive selection in genes encoding antiviral responses, components of the Toll and JAK/STAT pathways, and serine protease inhibitors in both social and solitary bees. Finally, we detect many genes across pathways that differ in selection between bumblebees and honeybees, or between the social and solitary clades.
Conclusions: The similarity in immune complement across a gradient of sociality suggests that a reduced immune repertoire predates the evolution of sociality in bees. The differences in selection on immune genes likely reflect divergent pressures exerted by parasites across social contexts.
Figures





Comment in
-
The making of eusociality: insights from two bumblebee genomes.Genome Biol. 2015 Apr 24;16(1):75. doi: 10.1186/s13059-015-0635-z. Genome Biol. 2015. PMID: 25908513 Free PMC article.
References
-
- Alexander RD. The evolution of social behavior. Annu Rev Ecol Systemat. 1974;5:325–83. doi: 10.1146/annurev.es.05.110174.001545. - DOI
-
- Foster SA. Group foraging by a coral-reef fish - a mechanism for gaining access to defended resources. Anim Behav. 1985;33:782–92. doi: 10.1016/S0003-3472(85)80011-7. - DOI
-
- Wilson EO. Success and dominance in ecosystems: the case of the social insects. Ecology Institute Oldendorf/Luhe: Oldendorf, Germany; 1990.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

