Molecular stratification of metastatic melanoma using gene expression profiling: Prediction of survival outcome and benefit from molecular targeted therapy
- PMID: 25909218
- PMCID: PMC4494939
- DOI: 10.18632/oncotarget.3655
Molecular stratification of metastatic melanoma using gene expression profiling: Prediction of survival outcome and benefit from molecular targeted therapy
Abstract
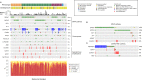
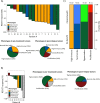
Melanoma is currently divided on a genetic level according to mutational status. However, this classification does not optimally predict prognosis. In prior studies, we have defined gene expression phenotypes (high-immune, pigmentation, proliferative and normal-like), which are predictive of survival outcome as well as informative of biology. Herein, we employed a population-based metastatic melanoma cohort and external cohorts to determine the prognostic and predictive significance of the gene expression phenotypes. We performed expression profiling on 214 cutaneous melanoma tumors and found an increased risk of developing distant metastases in the pigmentation (HR, 1.9; 95% CI, 1.05-3.28; P=0.03) and proliferative (HR, 2.8; 95% CI, 1.43-5.57; P=0.003) groups as compared to the high-immune response group. Further genetic characterization of melanomas using targeted deep-sequencing revealed similar mutational patterns across these phenotypes. We also used publicly available expression profiling data from melanoma patients treated with targeted or vaccine therapy in order to determine if our signatures predicted therapeutic response. In patients receiving targeted therapy, melanomas resistant to targeted therapy were enriched in the MITF-low proliferative subtype as compared to pre-treatment biopsies (P=0.02). In summary, the melanoma gene expression phenotypes are highly predictive of survival outcome and can further help to discriminate patients responding to targeted therapy.
Keywords: BRAF; BRAF inhibitor; gene expression; melanoma; mutation.
Conflict of interest statement
There is no conflict of interest that we should disclose.
Figures


References
-
- Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG, Ding S, Eggermont AM, Flaherty KT, Gimotty PA, Kirkwood JM, McMasters KM, Mihm MC, Jr, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199–6206. - PMC - PubMed
-
- Thompson JF, Scolyer RA, Kefford RF. Cutaneous melanoma. Lancet. 2005;365:687–701. - PubMed
-
- Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, Avril MF, Ortiz Romero PL, Robert T, Balacescu O, Eggermont AM, Lenoir G, Sarasin A, Tursz T, van den Oord JJ, Spatz A. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98:472–482. - PubMed
-
- Mann GJ, Pupo GM, Campain AE, Carter CD, Schramm SJ, Pianova S, Gerega SK, De Silva C, Lai K, Wilmott JS, Synnott M, Hersey P, Kefford RF, Thompson JF, Yang YH, Scolyer RA. BRAF mutation, NRAS mutation, and the absence of an immune-related expressed gene profile predict poor outcome in patients with stage III melanoma. J Invest Dermatol. 2013;133:509–517. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

