Genetic Variation along the Histamine Pathway in Children with Allergic versus Nonallergic Asthma
- PMID: 25909280
- PMCID: PMC4742940
- DOI: 10.1165/rcmb.2014-0493OC
Genetic Variation along the Histamine Pathway in Children with Allergic versus Nonallergic Asthma
Abstract
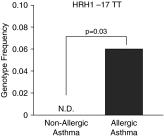
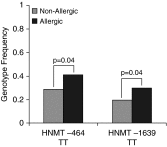
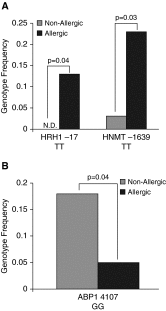
Histamine is an important mediator in the pathogenesis of asthma. Variation in genes along the histamine production, response, and degradation pathway may be important in predicting response to antihistamines. We hypothesize that differences exist among single-nucleotide polymorphisms (SNPs) in genes of the histamine pathway between children with allergic versus nonallergic asthma. Children (7-18 yr of age; n = 202) with asthma were classified as allergic or nonallergic based on allergy skin testing. Genotyping was performed to detect known SNPs (n = 10) among genes (HDC, HNMT, ABP1, HRH1, and HRH4) within the histamine pathway. Chi square tests and Cochran-Armitage Trend were used to identify associations between genetic variants and allergic or nonallergic asthma. Significance was determined by P < 0.05 and false-positive report probability. After correction for race differences in genotype were observed, HRH1-17 TT (6% allergic versus 0% nonallergic; P = 0.04), HNMT-464 TT (41% allergic versus 29% nonallergic; P = 0.04), and HNMT-1639 TT (30% allergic versus 20% nonallergic; P = 0.04) were overrepresented among children with allergic asthma. Genotype differences specifically among the African-American children were also observed: HRH1-17 TT (13% allergic versus 0% nonallergic; P = 0.04) and HNMT-1639 TT (23% allergic versus 3% nonallergic; P = 0.03) genotypes were overrepresented among African-American children with allergic asthma. Our study suggests that genetic variation within the histamine pathway may be associated with an allergic versus nonallergic asthma phenotype. Further studies are needed to determine the functional significance of identified SNPs and their impact on antihistamine response in patients with asthma and allergic disease.
Keywords: asthma; genetics; histamine.
Figures
References
-
- National Heart, Lung, and Blood Institute. Expert Panel Report 3 (EPR3): Guidelines for the diagnosis and management of asthma 2007 2007. [updated 2012 Apr; accessed 2014 Mar 1]Available from: http://www.nhlbi.nih.gov/guidelines/asthma/asthgdln.htm
-
- Barnes PJ. Pathophysiology of allergic inflammation. Immunol Rev. 2011;242:31–50. - PubMed
-
- Bourdin A, Gras D, Vachier I, Chanez P. Upper airway x 1: allergic rhinitis and asthma: united disease through epithelial cells. Thorax. 2009;64:999–1004. - PubMed
-
- Maintz L, Novak N. Histamine and histamine intolerance. Am J Clin Nutr. 2007;85:1185–1196. - PubMed
-
- García-Martín E, Ayuso P, Martínez C, Blanca M, Agúndez JA. Histamine pharmacogenomics. Pharmacogenomics. 2009;10:867–883. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

