Characterization of chromosomal and megaplasmid partitioning loci in Thermus thermophilus HB27
- PMID: 25909452
- PMCID: PMC4409726
- DOI: 10.1186/s12864-015-1523-3
Characterization of chromosomal and megaplasmid partitioning loci in Thermus thermophilus HB27
Abstract
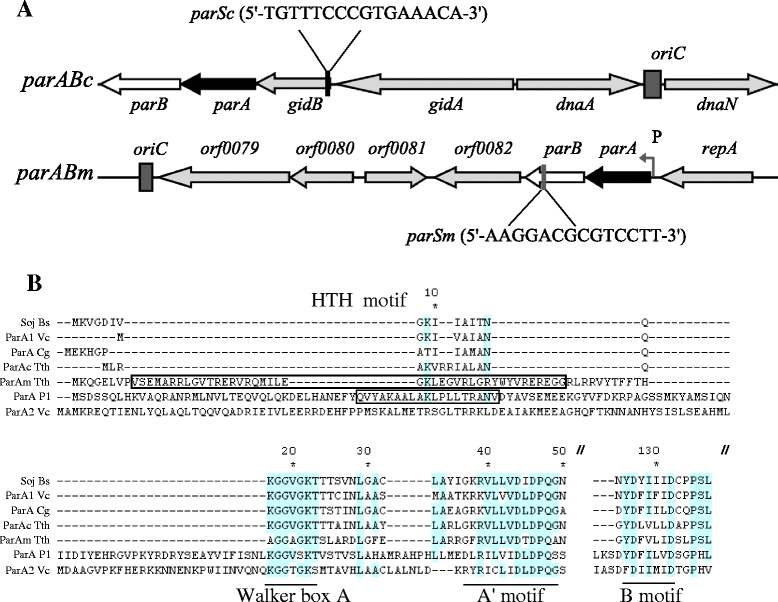
Background: In low-copy-number plasmids, the partitioning loci (par) act to ensure proper plasmid segregation and copy number maintenance in the daughter cells. In many bacterial species, par gene homologues are encoded on the chromosome, but their function is much less understood. In the two-replicon, polyploid genome of the hyperthermophilic bacterium Thermus thermophilus, both the chromosome and the megaplasmid encode par gene homologues (parABc and parABm, respectively). The mode of partitioning of the two replicons and the role of the two Par systems in the replication, segregation and maintenance of the genome copies are completely unknown in this organism.
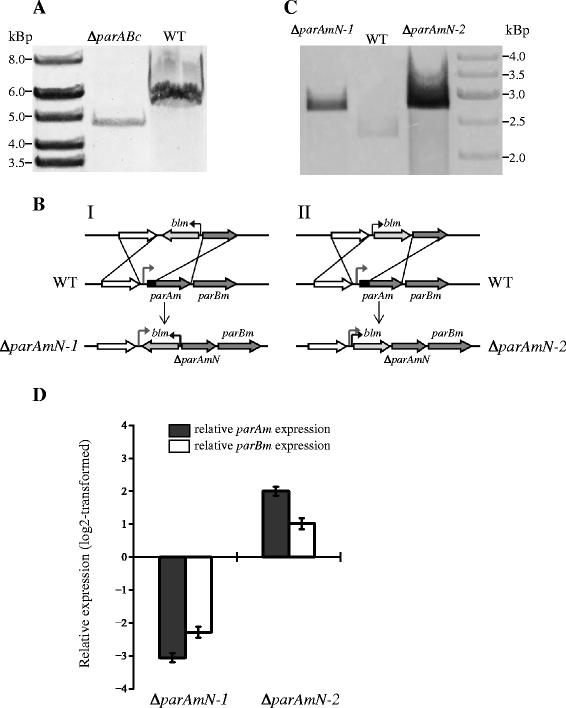
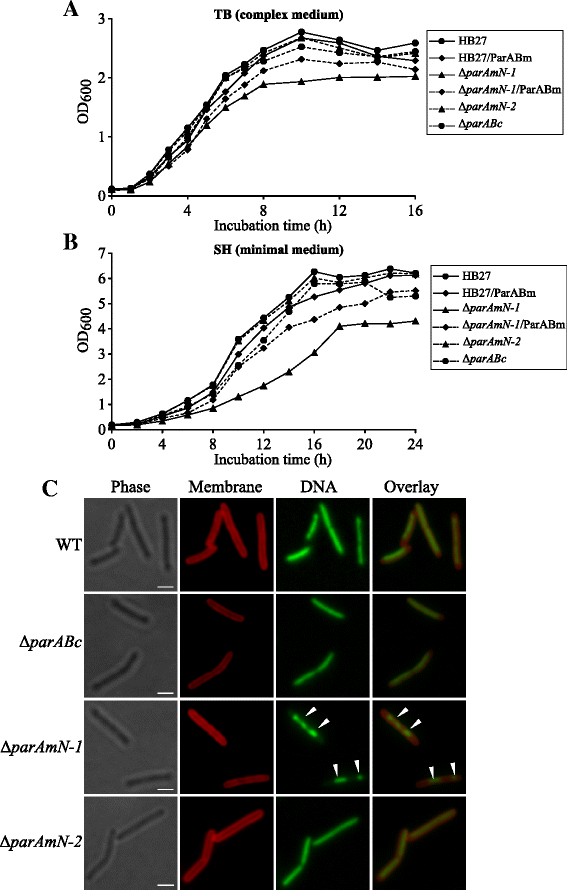
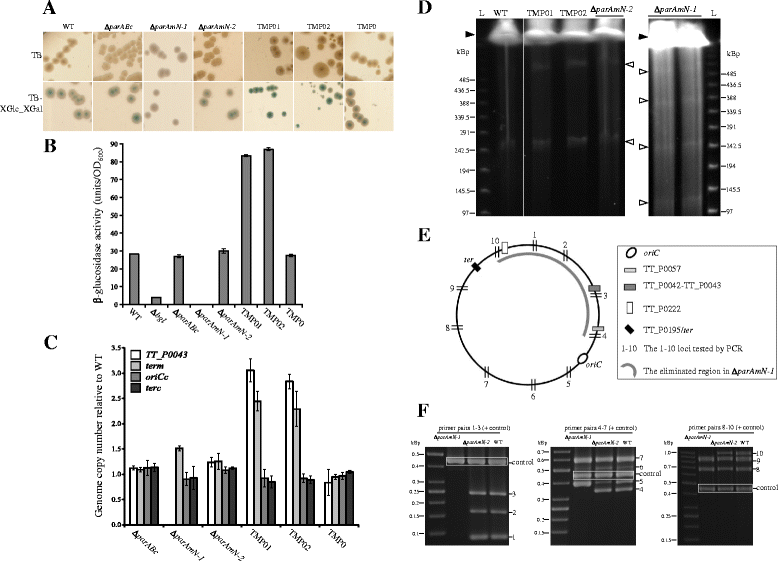
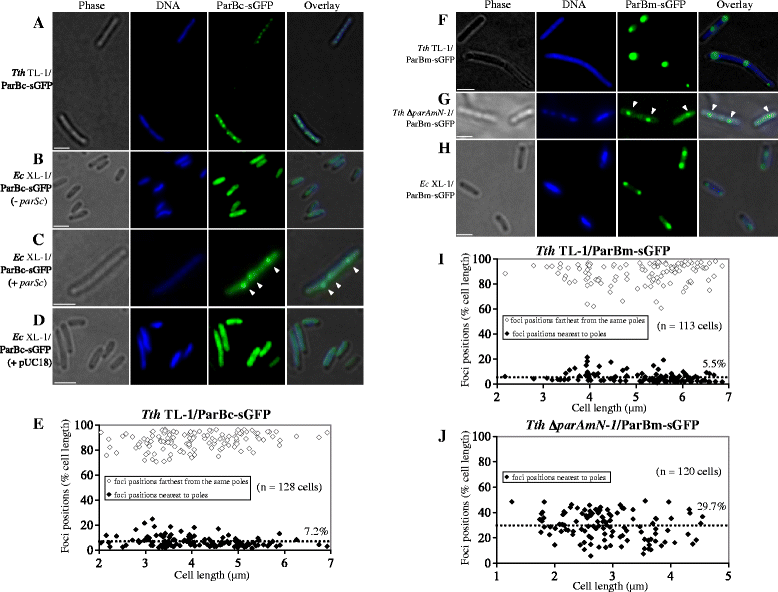
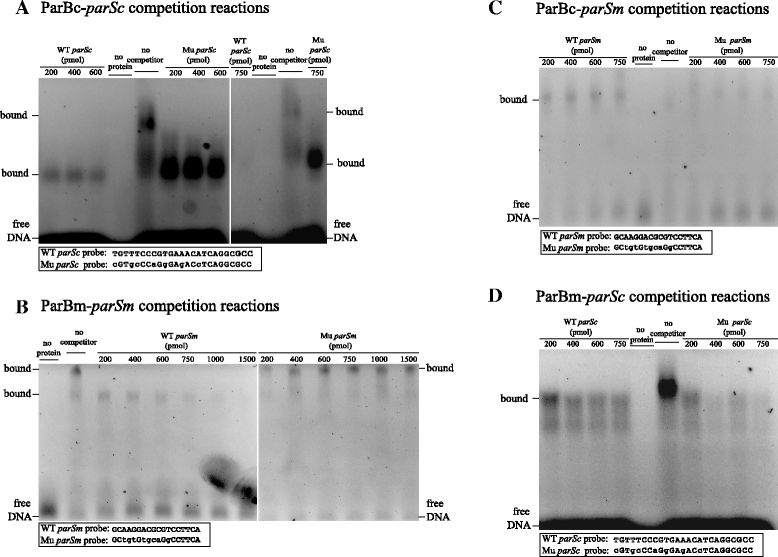
Results: We generated a series of chromosomal and megaplasmid par mutants and sGFP reporter strains and analyzed them with respect to DNA segregation defects, genome copy number and replication origin localization. We show that the two ParB proteins specifically bind their cognate centromere-like sequences parS, and that both ParB-parS complexes localize at the cell poles. Deletion of the chromosomal parAB genes did not apparently affect the cell growth, the frequency of cells with aberrant nucleoids, or the chromosome and megaplasmid replication. In contrast, deletion of the megaplasmid parAB operon or of the parB gene was not possible, indicating essentiality of the megaplasmid-encoded Par system. A mutant expressing lower amounts of ParABm showed growth defects, a high frequency of cells with irregular nucleoids and a loss of a large portion of the megaplasmid. The truncated megaplasmid could not be partitioned appropriately, as interlinked megaplasmid molecules (catenenes) could be detected, and the ParBm-parSm complexes in this mutant lost their polar localization.
Conclusions: We show that in T. thermophilus the chromosomal par locus is not required for either the chromosomal or megaplasmid bulk DNA replication and segregation. In contrast, the megaplasmid Par system of T. thermophilus is needed for the proper replication and segregation of the megaplasmid, and is essential for its maintenance. The two Par sets in T. thermophilus appear to function in a replicon-specific manner. To our knowledge, this is the first analysis of Par systems in a polyploid bacterium.
Figures
Similar articles
-
Random Chromosome Partitioning in the Polyploid Bacterium Thermus thermophilus HB27.G3 (Bethesda). 2019 Apr 9;9(4):1249-1261. doi: 10.1534/g3.119.400086. G3 (Bethesda). 2019. PMID: 30792193 Free PMC article.
-
The parABSm system is involved in megaplasmid partitioning and genome integrity maintenance in Thermus thermophilus.G3 (Bethesda). 2023 Apr 11;13(4):jkad038. doi: 10.1093/g3journal/jkad038. G3 (Bethesda). 2023. PMID: 36786449 Free PMC article.
-
Prokaryotic ParA-ParB-parS system links bacterial chromosome segregation with the cell cycle.Plasmid. 2012 Jan;67(1):1-14. doi: 10.1016/j.plasmid.2011.08.003. Epub 2011 Sep 6. Plasmid. 2012. PMID: 21924286 Review.
-
An extreme thermophile, Thermus thermophilus, is a polyploid bacterium.J Bacteriol. 2010 Oct;192(20):5499-505. doi: 10.1128/JB.00662-10. Epub 2010 Aug 20. J Bacteriol. 2010. PMID: 20729360 Free PMC article.
-
Plasmid and chromosome partitioning: surprises from phylogeny.Mol Microbiol. 2000 Aug;37(3):455-66. doi: 10.1046/j.1365-2958.2000.01975.x. Mol Microbiol. 2000. PMID: 10931339 Review.
Cited by
-
Random Chromosome Partitioning in the Polyploid Bacterium Thermus thermophilus HB27.G3 (Bethesda). 2019 Apr 9;9(4):1249-1261. doi: 10.1534/g3.119.400086. G3 (Bethesda). 2019. PMID: 30792193 Free PMC article.
-
Tracking pairwise genomic loci by the ParB-ParS and Noc-NBS systems in living cells.Nucleic Acids Res. 2024 May 22;52(9):4922-4934. doi: 10.1093/nar/gkae134. Nucleic Acids Res. 2024. PMID: 38412314 Free PMC article.
-
A type III-associated Cas6 functions as a negative regulator of type I-B CRISPR-Cas system in Thermus thermophilus.Commun Biol. 2025 May 23;8(1):793. doi: 10.1038/s42003-025-08223-4. Commun Biol. 2025. PMID: 40410287 Free PMC article.
-
The Divided Bacterial Genome: Structure, Function, and Evolution.Microbiol Mol Biol Rev. 2017 Aug 9;81(3):e00019-17. doi: 10.1128/MMBR.00019-17. Print 2017 Sep. Microbiol Mol Biol Rev. 2017. PMID: 28794225 Free PMC article. Review.
-
Nonequilibrium polysome dynamics promote chromosome segregation and its coupling to cell growth in Escherichia coli.Elife. 2025 Jun 24;14:RP104276. doi: 10.7554/eLife.104276. Elife. 2025. PMID: 40552714 Free PMC article.
References
-
- Webb CD, Graumann PL, Kahana JA, Teleman AA, Silver PA, Losick R. Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol Microbiol. 1998;28:883–92. doi: 10.1046/j.1365-2958.1998.00808.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

