Cell, isoform, and environment factors shape gradients and modulate chemotaxis
- PMID: 25909600
- PMCID: PMC4409393
- DOI: 10.1371/journal.pone.0123450
Cell, isoform, and environment factors shape gradients and modulate chemotaxis
Erratum in
-
Correction: Cell, Isoform, and Environment Factors Shape Gradients and Modulate Chemotaxis.PLoS One. 2017 Mar 14;12(3):e0174189. doi: 10.1371/journal.pone.0174189. eCollection 2017. PLoS One. 2017. PMID: 28291822 Free PMC article.
Abstract
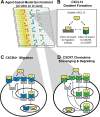
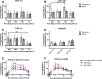
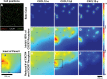
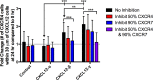
Chemokine gradient formation requires multiple processes that include ligand secretion and diffusion, receptor binding and internalization, and immobilization of ligand to surfaces. To understand how these events dynamically shape gradients and influence ensuing cell chemotaxis, we built a multi-scale hybrid agent-based model linking gradient formation, cell responses, and receptor-level information. The CXCL12/CXCR4/CXCR7 signaling axis is highly implicated in metastasis of many cancers. We model CXCL12 gradient formation as it is impacted by CXCR4 and CXCR7, with particular focus on the three most highly expressed isoforms of CXCL12. We trained and validated our model using data from an in vitro microfluidic source-sink device. Our simulations demonstrate how isoform differences on the molecular level affect gradient formation and cell responses. We determine that ligand properties specific to CXCL12 isoforms (binding to the migration surface and to CXCR4) significantly impact migration and explain differences in in vitro chemotaxis data. We extend our model to analyze CXCL12 gradient formation in a tumor environment and find that short distance, steep gradients characteristic of the CXCL12-γ isoform are effective at driving chemotaxis. We highlight the importance of CXCL12-γ in cancer cell migration: its high effective affinity for both extracellular surface sites and CXCR4 strongly promote CXCR4+ cell migration. CXCL12-γ is also more difficult to inhibit, and we predict that co-inhibition of CXCR4 and CXCR7 is necessary to effectively hinder CXCL12-γ-induced migration. These findings support the growing importance of understanding differences in protein isoforms, and in particular their implications for cancer treatment.
Conflict of interest statement
Figures


References
-
- Cavnar SP, Ray P, Moudgil P, Chang SL, Luker KE, Linderman JJ, et al. Microfluidic source-sink model reveals effects of biophysically distinct CXCL12 isoforms in breast cancer chemotaxis. Integrative biology: quantitative biosciences from nano to macro. 2014;6(5):564–76. Epub 2014/03/29. 10.1039/c4ib00015c PubMed . - DOI - PMC - PubMed
-
- Torisawa YS, Mosadegh B, Bersano-Begey T, Steele JM, Luker KE, Luker GD, et al. Microfluidic platform for chemotaxis in gradients formed by CXCL12 source-sink cells. Integrative biology: quantitative biosciences from nano to macro. 2010;2(11–12):680–6. Epub 2010/09/28. 10.1039/c0ib00041h PubMed . - DOI - PMC - PubMed
-
- Buckley CD, Amft N, Bradfield PF, Pilling D, Ross E, Arenzana-Seisdedos F, et al. Persistent induction of the chemokine receptor CXCR4 by TGF-beta 1 on synovial T cells contributes to their accumulation within the rheumatoid synovium. J Immunol. 2000;165(6):3423–9. Epub 2000/09/07. PubMed . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01CA136553/CA/NCI NIH HHS/United States
- R21CA182333/CA/NCI NIH HHS/United States
- R01CA142750/CA/NCI NIH HHS/United States
- R01 CA142750/CA/NCI NIH HHS/United States
- R01 CA136553/CA/NCI NIH HHS/United States
- P50CA093990/CA/NCI NIH HHS/United States
- T32 CA140044/CA/NCI NIH HHS/United States
- R21 CA182333/CA/NCI NIH HHS/United States
- R01 GM096040/GM/NIGMS NIH HHS/United States
- R01 EB012579/EB/NIBIB NIH HHS/United States
- EB012579/EB/NIBIB NIH HHS/United States
- P50 CA093990/CA/NCI NIH HHS/United States
- GM096040/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

