Diverse Phenotypes and Specific Transcription Patterns in Twenty Mouse Lines with Ablated LincRNAs
- PMID: 25909911
- PMCID: PMC4409293
- DOI: 10.1371/journal.pone.0125522
Diverse Phenotypes and Specific Transcription Patterns in Twenty Mouse Lines with Ablated LincRNAs
Abstract
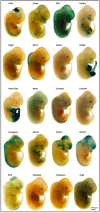
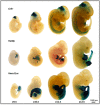
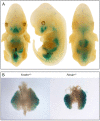
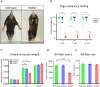
In a survey of 20 knockout mouse lines designed to examine the biological functions of large intergenic non-coding RNAs (lincRNAs), we have found a variety of phenotypes, ranging from perinatal lethality to defects associated with premature aging and morphological and functional abnormalities in the lungs, skeleton, and muscle. Each mutant allele carried a lacZ reporter whose expression profile highlighted a wide spectrum of spatiotemporal and tissue-specific transcription patterns in embryos and adults that informed our phenotypic analyses and will serve as a guide for future investigations of these genes. Our study shows that lincRNAs are a new class of encoded molecules that, like proteins, serve essential and important functional roles in embryonic development, physiology, and homeostasis of a broad array of tissues and organs in mammals.
Conflict of interest statement
Figures



References
-
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309: 1559–1563. - PubMed
-
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316: 1484–1488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

