Widespread macromolecular interaction perturbations in human genetic disorders
- PMID: 25910212
- PMCID: PMC4441215
- DOI: 10.1016/j.cell.2015.04.013
Widespread macromolecular interaction perturbations in human genetic disorders
Abstract
How disease-associated mutations impair protein activities in the context of biological networks remains mostly undetermined. Although a few renowned alleles are well characterized, functional information is missing for over 100,000 disease-associated variants. Here we functionally profile several thousand missense mutations across a spectrum of Mendelian disorders using various interaction assays. The majority of disease-associated alleles exhibit wild-type chaperone binding profiles, suggesting they preserve protein folding or stability. While common variants from healthy individuals rarely affect interactions, two-thirds of disease-associated alleles perturb protein-protein interactions, with half corresponding to "edgetic" alleles affecting only a subset of interactions while leaving most other interactions unperturbed. With transcription factors, many alleles that leave protein-protein interactions intact affect DNA binding. Different mutations in the same gene leading to different interaction profiles often result in distinct disease phenotypes. Thus disease-associated alleles that perturb distinct protein activities rather than grossly affecting folding and stability are relatively widespread.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
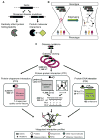
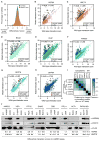
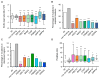


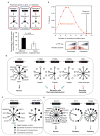
Comment in
-
Disease genetics: Network effects of disease mutations.Nat Rev Genet. 2015 Jun;16(6):317. doi: 10.1038/nrg3957. Nat Rev Genet. 2015. PMID: 25982168 No abstract available.
References
-
- Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM) Hum Mutat. 2011;32:564–567. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

