Human gene-centered transcription factor networks for enhancers and disease variants
- PMID: 25910213
- PMCID: PMC4409666
- DOI: 10.1016/j.cell.2015.03.003
Human gene-centered transcription factor networks for enhancers and disease variants
Abstract
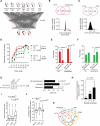
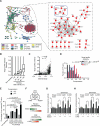
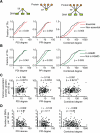
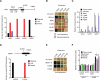
Gene regulatory networks (GRNs) comprising interactions between transcription factors (TFs) and regulatory loci control development and physiology. Numerous disease-associated mutations have been identified, the vast majority residing in non-coding regions of the genome. As current GRN mapping methods test one TF at a time and require the use of cells harboring the mutation(s) of interest, they are not suitable to identify TFs that bind to wild-type and mutant loci. Here, we use gene-centered yeast one-hybrid (eY1H) assays to interrogate binding of 1,086 human TFs to 246 enhancers, as well as to 109 non-coding disease mutations. We detect both loss and gain of TF interactions with mutant loci that are concordant with target gene expression changes. This work establishes eY1H assays as a powerful addition to the toolkit of mapping human GRNs and for the high-throughput characterization of genomic variants that are rapidly being identified by genome-wide association studies.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

