Gemin5 Binds to the Survival Motor Neuron mRNA to Regulate SMN Expression
- PMID: 25911097
- PMCID: PMC4505476
- DOI: 10.1074/jbc.M115.646257
Gemin5 Binds to the Survival Motor Neuron mRNA to Regulate SMN Expression
Abstract
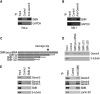
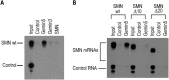
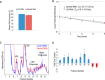
Reduced expression of SMN causes spinal muscular atrophy, a severe neurodegenerative disease. Despite the importance of maintaining SMN levels, relatively little is known about the mechanisms by which SMN levels are regulated. We show here that Gemin5, the snRNA-binding protein of the SMN complex, binds directly to the SMN mRNA and regulates SMN expression. Gemin5 binds with high specificity, both in vitro and in vivo, to sequence and structural elements in the SMN mRNA 3'-untranslated region that are reminiscent of the snRNP code to which Gemin5 binds on snRNAs. Reduction of Gemin5 redistributes the SMN mRNA from heavy polysomes to lighter polysomes and monosomes, suggesting that Gemin5 functions as an activator of SMN translation. SMN protein is not stoichiometrically present on the SMN mRNA with Gemin5, but the mRNA-binding activity of Gemin5 is dependent on SMN levels, providing a feedback mechanism for SMN to regulate its own expression via Gemin5. This work both reveals a new autoregulatory pathway governing SMN expression, and identifies a new mechanism through which SMN can modulate specific mRNA expression via Gemin5.
Keywords: Gemin5; RNA-binding protein; SMA; SMN; mRNA; neurodegenerative disease; post-transcriptional regulation; spinal muscular atrophy; survival motor neuron; translation.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

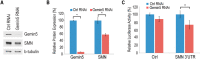
References
-
- Eggert C., Chari A., Laggerbauer B., Fischer U. (2006) Spinal muscular atrophy: the RNP connection. Trends Mol. Med. 12, 113–121 - PubMed
-
- Fischer U., Englbrecht C., Chari A. (2011) Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip. Rev. RNA 2, 718–731 - PubMed
-
- Liu Q., Fischer U., Wang F., Dreyfuss G. (1997) The Spinal Muscular Atrophy Disease Gene Product, SMN, and Its Associated Protein SIP1 Are in a Complex with Spliceosomal snRNP Proteins. Cell 90, 1013–1021 - PubMed
-
- Meister G., Eggert C., Fischer U. (2002) SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 12, 472–478 - PubMed
-
- Paushkin S., Gubitz A. K., Massenet S., Dreyfuss G. (2002) The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 14, 305–312 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

