The V4 and V5 Variable Loops of HIV-1 Envelope Glycoprotein Are Tolerant to Insertion of Green Fluorescent Protein and Are Useful Targets for Labeling
- PMID: 25911103
- PMCID: PMC4463467
- DOI: 10.1074/jbc.M114.628610
The V4 and V5 Variable Loops of HIV-1 Envelope Glycoprotein Are Tolerant to Insertion of Green Fluorescent Protein and Are Useful Targets for Labeling
Abstract
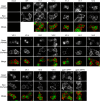
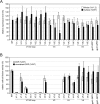
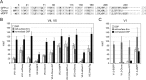
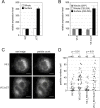
The mature human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) comprises the non-covalently associated gp120 and gp41 subunits generated from the gp160 precursor. Recent structural analyses have provided quaternary structural models for gp120/gp41 trimers, including the variable loops (V1-V5) of gp120. In these models, the V3 loop is located under V1/V2 at the apical center of the Env trimer, and the V4 and V5 loops project outward from the trimeric protomers. In addition, the V4 and V5 loops are predicted to have less movement upon receptor binding during membrane fusion events. We performed insertional mutagenesis using a GFP variant, GFPOPT, placed into the variable loops of HXB2 gp120. This allowed us to evaluate the current structural models and to simultaneously generate a GFP-tagged HIV-1 Env, which was useful for image analyses. All GFP-inserted mutants showed similar levels of whole-cell expression, although certain mutants, particularly V3 mutants, showed lower levels of cell surface expression. Functional evaluation of their fusogenicities in cell-cell and virus-like particle-cell fusion assays revealed that V3 was the most sensitive to the insertion and that the V1/V2 loops were less sensitive than V3. The V4 and V5 loops were the most tolerant to insertion, and certain tag proteins other than GFPOPT could also be inserted without functional consequences. Our results support the current structural models and provide a GFPOPT-tagged Env construct for imaging studies.
Keywords: envelope glycoprotein; human immunodeficiency virus (HIV); imaging; membrane fusion; mutagenesis; virus entry.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Moulard M., Decroly E. (2000) Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta 1469, 121–132 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

