Tracing the Evolutionary History of Inositol, 1, 4, 5-Trisphosphate Receptor: Insights from Analyses of Capsaspora owczarzaki Ca2+ Release Channel Orthologs
- PMID: 25911230
- PMCID: PMC4540961
- DOI: 10.1093/molbev/msv098
Tracing the Evolutionary History of Inositol, 1, 4, 5-Trisphosphate Receptor: Insights from Analyses of Capsaspora owczarzaki Ca2+ Release Channel Orthologs
Abstract
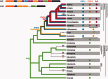
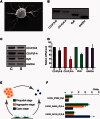
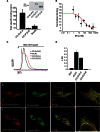
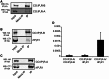
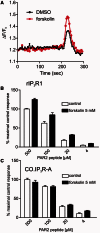
Cellular Ca(2+) homeostasis is tightly regulated and is pivotal to life. Inositol 1,4,5-trisphosphate receptors (IP3Rs) and ryanodine receptors (RyRs) are the major ion channels that regulate Ca(2+) release from intracellular stores. Although these channels have been extensively investigated in multicellular organisms, an appreciation of their evolution and the biology of orthologs in unicellular organisms is largely lacking. Extensive phylogenetic analyses reveal that the IP3R gene superfamily is ancient and diverged into two subfamilies, IP3R-A and IP3R-B/RyR, at the dawn of Opisthokonta. IP3R-B/RyR further diversified into IP3R-B and RyR at the stem of Filozoa. Subsequent evolution and speciation of Holozoa is associated with duplication of IP3R-A and RyR genes, and loss of IP3R-B in the vertebrate lineages. To gain insight into the properties of IP3R important for the challenges of multicellularity, the IP3R-A and IP3R-B family orthologs were cloned from Capsaspora owczarzaki, a close unicellular relative to Metazoa (designated as CO.IP3R-A and CO.IP3R-B). Both proteins were targeted to the endoplasmic reticulum. However, CO.IP3R-A, but strikingly not CO.IP3R-B, bound IP3, exhibited robust Ca(2+) release activity and associated with mammalian IP3Rs. These data indicate strongly that CO.IP3R-A as an exemplar of ancestral IP3R-A orthologs forms bona fide IP3-gated channels. Notably, however, CO.IP3R-A appears not to be regulated by Ca(2+), ATP or Protein kinase A-phosphorylation. Collectively, our findings explore the origin, conservation, and diversification of IP3R gene families and provide insight into the functionality of ancestral IP3Rs and the added specialization of these proteins in Metazoa.
Keywords: 4; 5-trisphosphate receptor; Capsaspora owczarzaki; calcium release channels; inositol 1.
© The Author 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

