Environmental control of branching in petunia
- PMID: 25911529
- PMCID: PMC4453797
- DOI: 10.1104/pp.15.00486
Environmental control of branching in petunia
Abstract
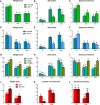
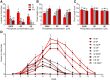
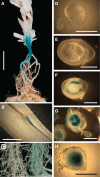
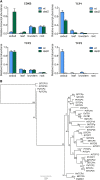
Plants alter their development in response to changes in their environment. This responsiveness has proven to be a successful evolutionary trait. Here, we tested the hypothesis that two key environmental factors, light and nutrition, are integrated within the axillary bud to promote or suppress the growth of the bud into a branch. Using petunia (Petunia hybrida) as a model for vegetative branching, we manipulated both light quality (as crowding and the red-to-far-red light ratio) and phosphate availability, such that the axillary bud at node 7 varied from deeply dormant to rapidly growing. In conjunction with the phenotypic characterization, we also monitored the state of the strigolactone (SL) pathway by quantifying SL-related gene transcripts. Mutants in the SL pathway inhibit but do not abolish the branching response to these environmental signals, and neither signal is dominant over the other, suggesting that the regulation of branching in response to the environment is complex. We have isolated three new putatively SL-related TCP (for Teosinte branched1, Cycloidia, and Proliferating cell factor) genes from petunia, and have identified that these TCP-type transcription factors may have roles in the SL signaling pathway both before and after the reception of the SL signal at the bud. We show that the abundance of the receptor transcript is regulated by light quality, such that axillary buds growing in added far-red light have greatly increased receptor transcript abundance. This suggests a mechanism whereby the impact of any SL signal reaching an axillary bud is modulated by the responsiveness of these cells to the signal.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures

Similar articles
-
Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia.Plant Physiol. 2009 Dec;151(4):1867-77. doi: 10.1104/pp.109.146720. Epub 2009 Oct 21. Plant Physiol. 2009. PMID: 19846541 Free PMC article.
-
Transcriptomic analysis implicates ABA signaling and carbon supply in the differential outgrowth of petunia axillary buds.BMC Plant Biol. 2023 Oct 10;23(1):482. doi: 10.1186/s12870-023-04505-3. BMC Plant Biol. 2023. PMID: 37814235 Free PMC article.
-
Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals.Plant Physiol. 2006 Mar;140(3):1109-17. doi: 10.1104/pp.105.074856. Epub 2006 Jan 27. Plant Physiol. 2006. PMID: 16443694 Free PMC article.
-
Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching.Plant Physiol. 2007 Feb;143(2):697-706. doi: 10.1104/pp.106.087957. Epub 2006 Dec 8. Plant Physiol. 2007. PMID: 17158589 Free PMC article.
-
Dynamics of strigolactone function and shoot branching responses in Pisum sativum.Mol Plant. 2013 Jan;6(1):128-40. doi: 10.1093/mp/sss131. Epub 2012 Dec 8. Mol Plant. 2013. PMID: 23220942
Cited by
-
BRANCHED1: A Key Hub of Shoot Branching.Front Plant Sci. 2019 Feb 12;10:76. doi: 10.3389/fpls.2019.00076. eCollection 2019. Front Plant Sci. 2019. PMID: 30809235 Free PMC article. Review.
-
Genome-Wide Identification, Characterization and Expression Analysis of TCP Transcription Factors in Petunia.Int J Mol Sci. 2020 Sep 9;21(18):6594. doi: 10.3390/ijms21186594. Int J Mol Sci. 2020. PMID: 32916908 Free PMC article.
-
Identification of Quantitative Trait Loci for Component Traits of Flowering Capacity Across Temperature in Petunia.G3 (Bethesda). 2019 Nov 5;9(11):3601-3610. doi: 10.1534/g3.119.400653. G3 (Bethesda). 2019. PMID: 31527047 Free PMC article.
-
Genome-Wide Identification and Characterization of the TCP Gene Family in Cucumber (Cucumis sativus L.) and Their Transcriptional Responses to Different Treatments.Genes (Basel). 2020 Nov 20;11(11):1379. doi: 10.3390/genes11111379. Genes (Basel). 2020. PMID: 33233827 Free PMC article.
-
Phloem Transport of the Receptor DWARF14 Protein Is Required for Full Function of Strigolactones.Plant Physiol. 2016 Nov;172(3):1844-1852. doi: 10.1104/pp.16.01212. Epub 2016 Sep 26. Plant Physiol. 2016. PMID: 27670819 Free PMC article.
References
-
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51: 1019–1029 - PubMed
-
- Arite T, Kameoka H, Kyozuka J (2012) Strigolactone positively controls crown root elongation in rice. J Plant Growth Regul 31: 165–172
-
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50: 1416–1424 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

