LATERAL ROOT PRIMORDIA 1 of maize acts as a transcriptional activator in auxin signalling downstream of the Aux/IAA gene rootless with undetectable meristem 1
- PMID: 25911745
- PMCID: PMC4473986
- DOI: 10.1093/jxb/erv187
LATERAL ROOT PRIMORDIA 1 of maize acts as a transcriptional activator in auxin signalling downstream of the Aux/IAA gene rootless with undetectable meristem 1
Abstract
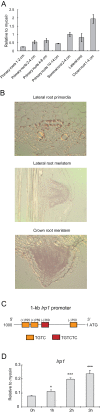
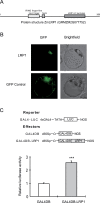
Only little is known about target genes of auxin signalling downstream of the Aux/IAA-ARF module. In the present study, it has been demonstrated that maize lateral root primordia 1 (lrp1) encodes a transcriptional activator that is directly regulated by the Aux/IAA protein ROOTLESS WITH UNDETECTABLE MERISTEM 1 (RUM1). Expression of lrp1 is confined to early root primordia and meristems and is auxin-inducible. Based on its primary protein structure, LRP1 is predicted to be a transcription factor. This notion is supported by exclusive LRP1 localization in the nucleus and its ability to activate downstream gene activity. Based on the observation that lrp1 transcription is completely repressed in the semi-dominant gain of function mutant rum1, it was demonstrated that the lrp1 promoter is a direct target of RUM1 proteins. Subsequently, promoter activation assays indicated that RUM1 represses the expression of a GFP reporter fused to the native promoter of lrp1. Constitutive repression of lrp1 in rum1 mutants is a consequence of the stability of mutated rum1 proteins which cannot be degraded by the proteasome and thus constitutively bind to the lrp1 promoter and repress transcription. Taken together, the repression of the transcriptional activator lrp1 by direct binding of RUM1 to its promoter, together with specific expression of lrp1 in root meristems, suggests a function in maize root development via the RUM1-dependent auxin signalling pathway.
Keywords: Aux/IAA; LATERAL ROOT PRIMORDIA 1 (LRP1); RUM1; crown root; differentiation zone; maize; transcriptional activator..
© The Author 2015. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures


References
-
- Abel S. 2007. Auxin is surfacing. ACS Chemical Biology 2, 380–384. - PubMed
-
- Benjamins R, Scheres B. 2008. Auxin: the looping star in plant development. Annual Reviews in Plant Biology 59, 443–465. - PubMed
-
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. 1993. Cellular organization of the Arabidopsis thaliana root. Development 119, 71–84. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

