Neuronal Activity Promotes Glioma Growth through Neuroligin-3 Secretion
- PMID: 25913192
- PMCID: PMC4447122
- DOI: 10.1016/j.cell.2015.04.012
Neuronal Activity Promotes Glioma Growth through Neuroligin-3 Secretion
Abstract
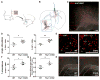
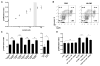
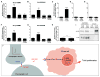
Active neurons exert a mitogenic effect on normal neural precursor and oligodendroglial precursor cells, the putative cellular origins of high-grade glioma (HGG). By using optogenetic control of cortical neuronal activity in a patient-derived pediatric glioblastoma xenograft model, we demonstrate that active neurons similarly promote HGG proliferation and growth in vivo. Conditioned medium from optogenetically stimulated cortical slices promoted proliferation of pediatric and adult patient-derived HGG cultures, indicating secretion of activity-regulated mitogen(s). The synaptic protein neuroligin-3 (NLGN3) was identified as the leading candidate mitogen, and soluble NLGN3 was sufficient and necessary to promote robust HGG cell proliferation. NLGN3 induced PI3K-mTOR pathway activity and feedforward expression of NLGN3 in glioma cells. NLGN3 expression levels in human HGG negatively correlated with patient overall survival. These findings indicate the important role of active neurons in the brain tumor microenvironment and identify secreted NLGN3 as an unexpected mechanism promoting neuronal activity-regulated cancer growth.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


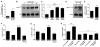
Comment in
-
Shedding light on glioma growth.Cell. 2015 May 7;161(4):704-6. doi: 10.1016/j.cell.2015.04.036. Cell. 2015. PMID: 25957677
-
A frightening thought: Neuronal activity enhances tumor growth.Cell Res. 2015 Aug;25(8):891-2. doi: 10.1038/cr.2015.65. Epub 2015 Jun 2. Cell Res. 2015. PMID: 26032268 Free PMC article.
-
How neuronal activity regulates glioma cell proliferation.Neuro Oncol. 2015 Dec;17(12):1543-4. doi: 10.1093/neuonc/nov188. Epub 2015 Sep 10. Neuro Oncol. 2015. PMID: 26359210 Free PMC article. No abstract available.
References
-
- Bergles DE, Roberts JD, Somogyi P, Jahr CE. Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature. 2000;405:187–191. - PubMed
-
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

