Structure of the Angiotensin receptor revealed by serial femtosecond crystallography
- PMID: 25913193
- PMCID: PMC4427029
- DOI: 10.1016/j.cell.2015.04.011
Structure of the Angiotensin receptor revealed by serial femtosecond crystallography
Abstract
Angiotensin II type 1 receptor (AT(1)R) is a G protein-coupled receptor that serves as a primary regulator for blood pressure maintenance. Although several anti-hypertensive drugs have been developed as AT(1)R blockers (ARBs), the structural basis for AT(1)R ligand-binding and regulation has remained elusive, mostly due to the difficulties of growing high-quality crystals for structure determination using synchrotron radiation. By applying the recently developed method of serial femtosecond crystallography at an X-ray free-electron laser, we successfully determined the room-temperature crystal structure of the human AT(1)R in complex with its selective antagonist ZD7155 at 2.9-Å resolution. The AT(1)R-ZD7155 complex structure revealed key structural features of AT(1)R and critical interactions for ZD7155 binding. Docking simulations of the clinically used ARBs into the AT(1)R structure further elucidated both the common and distinct binding modes for these anti-hypertensive drugs. Our results thereby provide fundamental insights into AT(1)R structure-function relationship and structure-based drug design.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures

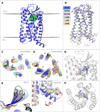
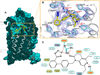
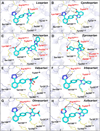


Comment in
-
Serial femtosecond crystallography datasets from G protein-coupled receptors.Sci Data. 2016 Aug 1;3:160057. doi: 10.1038/sdata.2016.57. Sci Data. 2016. PMID: 27479354 Free PMC article.
References
-
- Akazawa H, Yano M, Yabumoto C, Kudo-Sakamoto Y, Komuro I. Angiotensin II type 1 and type 2 receptor-induced cell signaling. Curr Pharm Des. 2013;19:2988–2995. - PubMed
-
- Balakumar P, Bishnoi HK, Mahadevan N. Telmisartan in the management of diabetic nephropathy: a contemporary view. Curr Diabetes Rev. 2012;8:183–190. - PubMed
-
- Balakumar P, Jagadeesh G. Structural determinants for binding, activation and functional selectivity of the AT1 receptor. J Mol Endocrinol. 2014;53:R71–R92. - PubMed
-
- Ballesteros JA, Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure–function relations in G-protein coupled receptors. Methods Neurosci. 1995;25:366–428.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R01HL57470/HL/NHLBI NIH HHS/United States
- T32 HL007914/HL/NHLBI NIH HHS/United States
- R01GM095583/GM/NIGMS NIH HHS/United States
- U54 GM094599/GM/NIGMS NIH HHS/United States
- R01 GM095583/GM/NIGMS NIH HHS/United States
- HL007914/HL/NHLBI NIH HHS/United States
- U54 GM094586/GM/NIGMS NIH HHS/United States
- U54 GM094618/GM/NIGMS NIH HHS/United States
- R01 HL057470/HL/NHLBI NIH HHS/United States
- R01 GM108635/GM/NIGMS NIH HHS/United States
- R01 HL115964/HL/NHLBI NIH HHS/United States
- U54GM094618/GM/NIGMS NIH HHS/United States
- P41 GM103393/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

