Deoxyribonucleic acid methylation profiling of single human blastocysts by methylated CpG-island amplification coupled with CpG-island microarray
- PMID: 25914096
- PMCID: PMC4449363
- DOI: 10.1016/j.fertnstert.2015.03.020
Deoxyribonucleic acid methylation profiling of single human blastocysts by methylated CpG-island amplification coupled with CpG-island microarray
Abstract
Objective: To study whether methylated CpG-island (CGI) amplification coupled with microarray (MCAM) can be used to generate DNA (deoxyribonucleic acid) methylation profiles from single human blastocysts.
Design: A pilot microarray study with methylated CpG-island amplification applied to human blastocyst genomic DNA and hybridized on CpG-island microarrays.
Setting: University research laboratory.
Patient(s): Five cryopreserved sibling 2-pronuclear zygotes that were surplus to requirements for clinical treatment by in vitro fertilization were donated with informed consent from a patient attending Bourn Hall Clinic, Cambridge, United Kingdom.
Intervention(s): None.
Main outcome measure(s): Successful generation of genome-wide DNA methylation profiles at CpG islands from individual human blastocysts, with common genomic regions of DNA methylation identified between embryos.
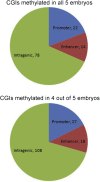
Result(s): Between 472 and 734 CpG islands were methylated in each blastocyst, with 121 CpG islands being commonly methylated in all 5 blastocysts. A further 159 CGIs were commonly methylated in 4 of the 5 tested blastocysts. Methylation was observed at a number of CGIs within imprinted-gene, differentially methylated regions (DMRs), including placental and preimplantation-specific DMRs.
Conclusion(s): The MCAM method is capable of providing comprehensive DNA methylation data in individual human blastocysts.
Keywords: CpG island; Preimplantation; blastocyst; epigenetic; methylation.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
High-throughput methylation profiling by MCA coupled to CpG island microarray.Genome Res. 2007 Oct;17(10):1529-36. doi: 10.1101/gr.6417007. Epub 2007 Sep 4. Genome Res. 2007. PMID: 17785535 Free PMC article.
-
Alterations in the sperm histone-retained epigenome are associated with unexplained male factor infertility and poor blastocyst development in donor oocyte IVF cycles.Hum Reprod. 2017 Dec 1;32(12):2443-2455. doi: 10.1093/humrep/dex317. Hum Reprod. 2017. PMID: 29087470
-
Dynamic CpG island methylation landscape in oocytes and preimplantation embryos.Nat Genet. 2011 Jun 26;43(8):811-4. doi: 10.1038/ng.864. Nat Genet. 2011. PMID: 21706000 Free PMC article.
-
Monitoring methylation changes in cancer.Adv Biochem Eng Biotechnol. 2007;104:1-11. doi: 10.1007/10_024. Adv Biochem Eng Biotechnol. 2007. PMID: 17290816 Review.
-
DNA methylation profiling using the methylated-CpG island recovery assay (MIRA).Methods. 2010 Nov;52(3):213-7. doi: 10.1016/j.ymeth.2010.03.004. Epub 2010 Mar 19. Methods. 2010. PMID: 20304072 Free PMC article. Review.
References
-
- Monk M., Boubelik M., Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. - PubMed
-
- Smallwood S.A., Kelsey G. De novo DNA methylation: a germ cell perspective. Trends Genet. 2012;28:33–42. - PubMed
-
- Lees-Murdock D.J., Walsh C.P. DNA methylation reprogramming in the germ line. Epigenetics. 2008;3:5–13. - PubMed
-
- Huntriss J., Picton H.M. Epigenetic consequences of assisted reproduction and infertility on the human preimplantation embryo. Hum Fertil (Camb) 2008;11:85–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

