DOCK 6: Impact of new features and current docking performance
- PMID: 25914306
- PMCID: PMC4469538
- DOI: 10.1002/jcc.23905
DOCK 6: Impact of new features and current docking performance
Abstract
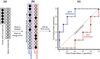
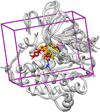
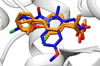
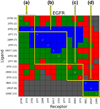
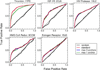
This manuscript presents the latest algorithmic and methodological developments to the structure-based design program DOCK 6.7 focused on an updated internal energy function, new anchor selection control, enhanced minimization options, a footprint similarity scoring function, a symmetry-corrected root-mean-square deviation algorithm, a database filter, and docking forensic tools. An important strategy during development involved use of three orthogonal metrics for assessment and validation: pose reproduction over a large database of 1043 protein-ligand complexes (SB2012 test set), cross-docking to 24 drug-target protein families, and database enrichment using large active and decoy datasets (Directory of Useful Decoys [DUD]-E test set) for five important proteins including HIV protease and IGF-1R. Relative to earlier versions, a key outcome of the work is a significant increase in pose reproduction success in going from DOCK 4.0.2 (51.4%) → 5.4 (65.2%) → 6.7 (73.3%) as a result of significant decreases in failure arising from both sampling 24.1% → 13.6% → 9.1% and scoring 24.4% → 21.1% → 17.5%. Companion cross-docking and enrichment studies with the new version highlight other strengths and remaining areas for improvement, especially for systems containing metal ions. The source code for DOCK 6.7 is available for download and free for academic users at http://dock.compbio.ucsf.edu/.
Keywords: DOCK; cross-docking; docking; enrichment; ligand flexibility; pose reproduction; virtual screening.
© 2015 Wiley Periodicals, Inc.
Figures












References
-
- Kuntz ID. Science. 1992;257:1078–1082. - PubMed
-
- Jorgensen WL. Science. 2004;303:1813–1818. - PubMed
-
- DesJarlais RL, Sheridan RP, Seibel GL, Dixon JS, Kuntz ID, Venkataraghavan R. J. Med. Chem. 1988;31:722–729. - PubMed
-
- Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. J. Mol. Biol. 1982;161:269–288. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

