Evaluation of Normalization and PCA on the Performance of Classifiers for Protein Crystallization Images
- PMID: 25914519
- PMCID: PMC4409005
- DOI: 10.1109/SECON.2014.6950744
Evaluation of Normalization and PCA on the Performance of Classifiers for Protein Crystallization Images
Abstract
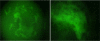
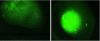
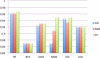
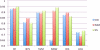
In this paper, we investigate the performance of classification of protein crystallization images captured during protein crystal growth process. We group protein crystallization images into 3 categories: noncrystals, likely leads (conditions that may yield formation of crystals) and crystals. In this research, we only consider the subcategories of noncrystal and likely leads protein crystallization images separately. We use 5 different classifiers to solve this problem and we applied some data preprocessing methods such as principal component analysis (PCA), min-max (MM) normalization and z-score (ZS) normalization methods to our datasets in order to evaluate their effects on classifiers for the noncrystal and likely leads datasets. We performed our experiments on 1606 noncrystal and 245 likely leads images independently. We had satisfactory results for both datasets. We reached 96.8% accuracy for noncrystal dataset and 94.8% accuracy for likely leads dataset. Our target is to investigate the best classifiers with optimal preprocessing techniques on both noncrystal and likely leads datasets.
Keywords: classification; normalization; principal component analysis; protein crystallization.
Figures
References
-
- Pan S, et al. Automated classification of protein crystallization images using support vector machines with scale-invariant texture and Gabor features. Acta Crystallographica Section D. 2006 Mar;62(3):271–279. - PubMed
-
- Rupp B, Wang J. Predictive Models for Protein Crystallization. Methods. 2004;34(3):390–407. - PubMed
-
- Bern M, Goldberg D, Stevens RC, Kuhn P. Automatic classification of protein crystallization images using a curve-tracking algorithm. Journal of Applied Crystallography. 2007 Apr;37(2):279–-287.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources