Ebola virus infection modeling and identifiability problems
- PMID: 25914675
- PMCID: PMC4391033
- DOI: 10.3389/fmicb.2015.00257
Ebola virus infection modeling and identifiability problems
Abstract
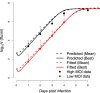
The recent outbreaks of Ebola virus (EBOV) infections have underlined the impact of the virus as a major threat for human health. Due to the high biosafety classification of EBOV (level 4), basic research is very limited. Therefore, the development of new avenues of thinking to advance quantitative comprehension of the virus and its interaction with the host cells is urgently needed to tackle this lethal disease. Mathematical modeling of the EBOV dynamics can be instrumental to interpret Ebola infection kinetics on quantitative grounds. To the best of our knowledge, a mathematical modeling approach to unravel the interaction between EBOV and the host cells is still missing. In this paper, a mathematical model based on differential equations is used to represent the basic interactions between EBOV and wild-type Vero cells in vitro. Parameter sets that represent infectivity of pathogens are estimated for EBOV infection and compared with influenza virus infection kinetics. The average infecting time of wild-type Vero cells by EBOV is slower than in influenza infection. Simulation results suggest that the slow infecting time of EBOV could be compensated by its efficient replication. This study reveals several identifiability problems and what kind of experiments are necessary to advance the quantification of EBOV infection. A first mathematical approach of EBOV dynamics and the estimation of standard parameters in viral infections kinetics is the key contribution of this work, paving the way for future modeling works on EBOV infection.
Keywords: EBOV; Ebola; identifiability; kinetics; mathematical modeling; viral dynamics.
Figures

Similar articles
-
Modeling Challenges of Ebola Virus-Host Dynamics during Infection and Treatment.Viruses. 2020 Jan 16;12(1):106. doi: 10.3390/v12010106. Viruses. 2020. PMID: 31963118 Free PMC article.
-
Enhancement of Ebola Virus Infection via Ficolin-1 Interaction with the Mucin Domain of GP Glycoprotein.J Virol. 2016 May 12;90(11):5256-5269. doi: 10.1128/JVI.00232-16. Print 2016 Jun 1. J Virol. 2016. PMID: 26984723 Free PMC article.
-
NK Cells Accumulate in Infected Tissues and Contribute to Pathogenicity of Ebola Virus in Mice.J Virol. 2019 May 1;93(10):e01703-18. doi: 10.1128/JVI.01703-18. Print 2019 May 15. J Virol. 2019. PMID: 30814283 Free PMC article.
-
Comprehensive Review on Ebola (EBOV) Virus: Future Prospects.Infect Disord Drug Targets. 2018;18(2):96-104. doi: 10.2174/1871526517666170817100828. Infect Disord Drug Targets. 2018. PMID: 28820067 Review.
-
Mathematical Analysis of Viral Replication Dynamics and Antiviral Treatment Strategies: From Basic Models to Age-Based Multi-Scale Modeling.Front Microbiol. 2018 Jul 11;9:1546. doi: 10.3389/fmicb.2018.01546. eCollection 2018. Front Microbiol. 2018. PMID: 30050523 Free PMC article. Review.
Cited by
-
Controlling of pandemic COVID-19 using optimal control theory.Results Phys. 2021 Jul;26:104311. doi: 10.1016/j.rinp.2021.104311. Epub 2021 May 19. Results Phys. 2021. PMID: 34094820 Free PMC article.
-
Characterization of SARS-CoV-2 dynamics in the host.Annu Rev Control. 2020;50:457-468. doi: 10.1016/j.arcontrol.2020.09.008. Epub 2020 Oct 6. Annu Rev Control. 2020. PMID: 33041634 Free PMC article.
-
Mechanisms of phosphatidylserine influence on viral production: A computational model of Ebola virus matrix protein assembly.J Biol Chem. 2022 Jul;298(7):102025. doi: 10.1016/j.jbc.2022.102025. Epub 2022 May 11. J Biol Chem. 2022. PMID: 35568195 Free PMC article.
-
Modeling Influenza Virus Infection: A Roadmap for Influenza Research.Viruses. 2015 Oct 12;7(10):5274-304. doi: 10.3390/v7102875. Viruses. 2015. PMID: 26473911 Free PMC article. Review.
-
Towards detection and diagnosis of Ebola virus disease at point-of-care.Biosens Bioelectron. 2016 Jan 15;75:254-72. doi: 10.1016/j.bios.2015.08.040. Epub 2015 Aug 20. Biosens Bioelectron. 2016. PMID: 26319169 Free PMC article. Review.
References
-
- Bowden D. (1983). Cell turnover in the lung. Am. Rev. Res. Dis. 128(2 Pt 2), S46–S48. - PubMed
-
- Brun R., Reichert P., Kuensch H. R. (2001). Practical identifiability analysis of large environmental simulation models. Water Res. Res. 37, 1015–1030 10.1029/2000WR900350 - DOI
-
- Carter J., Saunders V. (2013). Virology: Principle and Applications. Chichester, UK: John Wiley.
LinkOut - more resources
Full Text Sources
Other Literature Sources

