STIM1 Is a Novel Component of ER-Chlamydia trachomatis Inclusion Membrane Contact Sites
- PMID: 25915399
- PMCID: PMC4411163
- DOI: 10.1371/journal.pone.0125671
STIM1 Is a Novel Component of ER-Chlamydia trachomatis Inclusion Membrane Contact Sites
Abstract
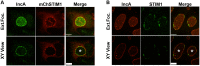
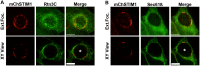
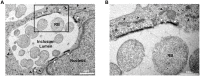
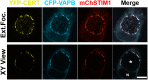
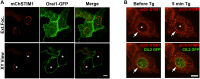
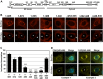
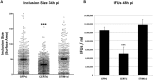
Productive developmental cycle of the obligate intracellular bacterial pathogen Chlamydia trachomatis depends on the interaction of the replicative vacuole, named the inclusion, with cellular organelles. We have recently reported the formation of ER-Inclusion membrane contact sites (MCSs), where the endoplasmic reticulum (ER) is in apposition to the inclusion membrane. These platforms contain the C. trachomatis inclusion membrane protein IncD, the mammalian ceramide transfer protein CERT and the ER resident proteins VAPA/B and were proposed to play a role in the non-vesicular trafficking of lipids to the inclusion. Here, we identify STIM1 as a novel component of ER-Inclusion MCSs. STIM1, an ER calcium (Ca2+) sensor that relocate to ER-Plasma Membrane (PM) MCSs upon Ca2+ store depletion, associated with C. trachomatis inclusion. STIM1, but not the general ER markers Rtn3C and Sec61ß, was enriched at the inclusion membrane. Ultra-structural studies demonstrated that STIM1 localized to ER-Inclusion MCSs. Time-course experiments showed that STIM1, CERT and VAPB co-localized throughout the developmental cycle. By contrast, Orai1, the PM Ca2+ channel that interacts with STIM1 at ER-PM MCSs, did not associate with C. trachomatis inclusion. Upon ER Ca2+ store depletion, a pool of STIM1 relocated to ER-PM MCSs, while the existing ER-Inclusion MCSs remained enriched in STIM1. Finally, we have identified the CAD domain, which mediates STIM1-Orai1 interaction, as the minimal domain required for STIM1 enrichment at ER-Inclusion MCSs. Altogether this study identifies STIM1 as a novel component of ER-C. trachomatis inclusion MCSs. We discuss the potential role(s) of STIM1 during the infection process.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

