Deubiquitinating enzymes regulate PARK2-mediated mitophagy
- PMID: 25915564
- PMCID: PMC4502823
- DOI: 10.1080/15548627.2015.1034408
Deubiquitinating enzymes regulate PARK2-mediated mitophagy
Abstract
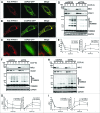
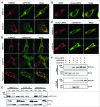
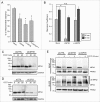
The selective degradation of mitochondria by the process of autophagy, termed mitophagy, is one of the major mechanisms of mitochondrial quality control. The best-studied mitophagy pathway is the one mediated by PINK1 and PARK2/Parkin. From recent studies it has become clear that ubiquitin-ligation plays a pivotal role and most of the focus has been on the role of ubiquitination of mitochondrial proteins in mitophagy. Even though ubiquitination is a reversible process, very little is known about the role of deubiquitinating enzymes (DUBs) in mitophagy. Here, we report that 2 mitochondrial DUBs, USP30 and USP35, regulate PARK2-mediated mitophagy. We show that USP30 and USP35 can delay PARK2-mediated mitophagy using a quantitative mitophagy assay. Furthermore, we show that USP30 delays mitophagy by delaying PARK2 recruitment to the mitochondria during mitophagy. USP35 does not delay PARK2 recruitment, suggesting that it regulates mitophagy through an alternative mechanism. Interestingly, USP35 only associates with polarized mitochondria, and rapidly translocates to the cytosol during CCCP-induced mitophagy. It is clear that PARK2-mediated mitophagy is regulated at many steps in this important quality control pathway. Taken together, these findings demonstrate an important role of mitochondrial-associated DUBs in mitophagy. Because defects in mitochondria quality control are implicated in many neurodegenerative disorders, our study provides clear rationales for the design and development of drugs for the therapeutic treatment of neurodegenerative diseases such as Parkinson and Alzheimer diseases.
Keywords: CCCP; Cer, cerulean; DMSO, dimethyl sulfoxide; DUB; DsRed, Discosoma sp. red fluorescent protein; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GFP, green fluorescent protein; HA, human influenza hemagglutinin; MFN2, mitofusin 2; MTS; OMM, outer mitochondrial membrane; PARK2; PARK2, parkin RBR E3 ubiquitin protein ligase; PINK1, PTEN-induced putative kinase 1; SYNJ2BP, synaptojanin 2 binding protein; TOMM20, translocase of outer mitochondrial membrane 20 homolog (yeast); USP, ubiquitin specific peptidase; USP30; USP35; autophagy; carbonyl cyanide m-chlorophenylhydrazone; deubiquitinating enzyme; deubiquitinating enzymes; l-USP35, long form of ubiquitin specific peptidase 35; mitochondrial dynamics; mitochondrial targeting sequence; mitophagy; neurodegenerative diseases; s-USP35, short form of ubiquitin specific peptidase 35; ubiquitin.
Figures

References
-
- Deas E, Wood NW, Plun-Favreau H. Mitophagy and Parkinson's disease: the PINK1-parkin link. Biochim Biophys Acta [Internet] 2011. [cited 2014 Apr 29]; 1813:623–33. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3925795&tool=p...; http://dx.doi.org/ 10.1016/j.bbamcr.2010.08.007 - DOI - PMC - PubMed
-
- Berman S, Pineda F, Hardwick J. Mitochondrial fission and fusion dynamics: the long and short of it. Cell Death Differ 2008; 15:1147–52; PMID:18437161; http://dx.doi.org/ 10.1038/cdd.2008.57 - DOI - PMC - PubMed
-
- Novak I. Mitophagy: a complex mechanism of mitochondrial removal. Antioxid Redox Signal [Internet] 2012. [cited 2012 Mar 23]; 0 Available from: http://www.ncbi.nlm.nih.gov/pubmed/22077334 - PubMed
-
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol [Internet] 2011. [cited 2010 Dec 22]; 12:9–14. Available from: http://www.nature.com/doifinder/10.1038/nrm3028; http://dx.doi.org/ 10.1038/nrm3028 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
