Enhanced Sensitivity for Detection of HIV-1 p24 Antigen by a Novel Nuclease-Linked Fluorescence Oligonucleotide Assay
- PMID: 25915630
- PMCID: PMC4410951
- DOI: 10.1371/journal.pone.0125701
Enhanced Sensitivity for Detection of HIV-1 p24 Antigen by a Novel Nuclease-Linked Fluorescence Oligonucleotide Assay
Abstract
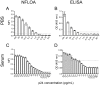
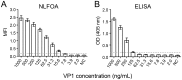
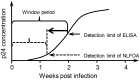
The relatively high detection limit of the Enzyme-linked immunosorbent assay (ELISA) prevents its application for detection of low concentrations of antigens. To increase the sensitivity for detection of HIV-1 p24 antigen, we developed a highly sensitive nuclease-linked fluorescence oligonucleotide assay (NLFOA). Two major improvements were incorporated in NLFOA to amplify antibody-antigen interaction signals and reduce the signal/noise ratio; a large number of nuclease molecules coupled to the gold nanoparticle/streptavidin complex and fluorescent signals generated from fluorescent-labeled oligonucleotides by the nuclease. The detection limit of p24 by NLFOA was 1 pg/mL, which was 10-fold more sensitive than the conventional ELISA (10 pg/mL). The specificity was 100% and the coefficient of variation (CV) was 7.8% at low p24 concentration (1.5 pg/mL) with various concentrations of spiked p24 in HIV-1 negative sera. Thus, NLFOA is highly sensitive, specific, reproducible and user-friendly. The more sensitive detection of low p24 concentrations in HIV-1-infected individuals by NLFOA could allow detection of HIV-1 infections that are missed by the conventional ELISA at the window period during acute infection to further reduce the risk for HIV-1 infection due to the undetected HIV-1 in the blood products. Moreover, NLFOA can be easily applied to more sensitive detection of other antigens.
Conflict of interest statement
Figures





References
-
- Lequin RM. Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). Clin Chem. 2005;51: 2415–2418. - PubMed
-
- Crowther J. Enzyme Linked Immunosorbent Assay (ELISA) In: Walker JM, Rapley R, editors. Molecular Biomethods Handbook. 2nd ed New York: Humana Press; 2008. p. 657–682.
-
- Meng Y, High K, Antonello J, Washabaugh MW, Zhao Q. Enhanced sensitivity and precision in an enzyme-linked immunosorbent assay with fluorogenic substrates compared with commonly used chromogenic substrates. Anal Biochem. 2005;345: 227–236. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

