Viral bimolecular fluorescence complementation: a novel tool to study intracellular vesicular trafficking pathways
- PMID: 25915798
- PMCID: PMC4411132
- DOI: 10.1371/journal.pone.0125619
Viral bimolecular fluorescence complementation: a novel tool to study intracellular vesicular trafficking pathways
Abstract
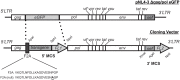
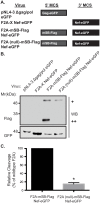
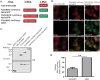
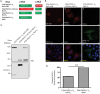
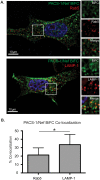
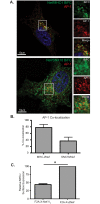
The Human Immunodeficiency Virus type 1 (HIV-1) accessory protein Nef interacts with a multitude of cellular proteins, manipulating the host membrane trafficking machinery to evade immune surveillance. Nef interactions have been analyzed using various in vitro assays, co-immunoprecipitation studies, and more recently mass spectrometry. However, these methods do not evaluate Nef interactions in the context of viral infection nor do they define the sub-cellular location of these interactions. In this report, we describe a novel bimolecular fluorescence complementation (BiFC) lentiviral expression tool, termed viral BiFC, to study Nef interactions with host cellular proteins in the context of viral infection. Using the F2A cleavage site from the foot and mouth disease virus we generated a viral BiFC expression vector capable of concurrent expression of Nef and host cellular proteins; PACS-1, MHC-I and SNX18. Our studies confirmed the interaction between Nef and PACS-1, a host membrane trafficking protein involved in Nef-mediated immune evasion, and demonstrated co-localization of this complex with LAMP-1 positive endolysosomal vesicles. Furthermore, we utilized viral BiFC to localize the Nef/MHC-I interaction to an AP-1 positive endosomal compartment. Finally, viral BiFC was observed between Nef and the membrane trafficking regulator SNX18. This novel demonstration of an association between Nef and SNX18 was localized to AP-1 positive vesicles. In summary, viral BiFC is a unique tool designed to analyze the interaction between Nef and host cellular proteins by mapping the sub-cellular locations of their interactions during viral infection.
Conflict of interest statement
Figures
References
-
- Kestler HW 3rd, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, et al. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65(4):651–62. Epub 1991/05/17. 0092-8674(91)90097-I [pii]. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

